Abstract
The IL12B gene encodes the common p40 subunit of IL-12 and IL-23, cytokines with key roles in Th1 and Th17 biology, respectively, and genetic variation in this region significantly influences risk of psoriasis. Here, we demonstrate that a psoriasis-associated risk haplotype at the IL12B locus leads to increased expression of IL12B by monocytes and correlated with increased serum levels of IL-12, IFN-γ and the IFN-γ induced chemokine, CXCL10. In contrast, serum IL-23 levels were decreased in risk carriers when compared with non-carriers. We further demonstrate that IL-12 is increased in psoriatic skin and that risk carriers manifest a skewing of the inflammatory network toward stronger IFN-γ responses. Taken together, our data demonstrate that the risk variant in IL12B associates with its increased expression and predisposes to stronger Th1 polarization through deviation of the local inflammatory environment toward increased IL-12/IFN-γ at the expense of IL-23/IL-17 responses.
INTRODUCTION
Psoriasis is one of the most common chronic inflammatory diseases affecting approximately 2–3% of Caucasians (1,2), but is found in all populations (1). The most common form, psoriasis vulgaris, is characterized by sharply demarcated red scaly plaques on the elbows, scalp and knees, and approximately 25% of patients go on to develop a debilitating inflammatory arthritis. It has been known for a long time that the predisposition to psoriasis is to a large extent genetic (2–4), and in recent years, considerable progress has been made in identifying these risk factors (4). One of these risk loci is proximal to the IL12B gene on chromosome 5q31.1–33.1 (5). In the initial report (5), two single nucleotide polymorphisms (SNPs), which are in linkage disequilibrium, were identified: rs3212227 located in the 3′UTR and rs6887695 located upstream of the IL12B gene.
The IL12B gene encodes the common p40 subunit of IL-12 (6) and IL-23 (7), key cytokines in Th1 and Th17 differentiation and function (8), and this gene's role in psoriasis pathogenesis is illustrated by the clinical efficacy observed with anti-p40 treatments (9,10). Although no studies have been published on the effect of the rs6887695 SNP, several studies have attempted to address the role of the 3′UTR SNP rs3212227 on IL-12p40 expression and secretion. Overall, the results from these studies have been inconsistent. Thus, whereas some have shown increased IL-12p40 expression (11) and secretion (12,13), others have shown decreased IL-12p70 production for the psoriasis-associated rs3212227 A allele (14).
In this paper, we report that the IL12B psoriasis risk haplotype defined by the G allele of rs6887695, and its associated A allele of rs3212227 [haplotype odds ratio of 1.52 (15)], leads to increased expression of the IL12p40 subunit by monocytes, the primary cellular source of IL-12 and IL-23 (16) and that this is further enhanced by IFN-γ stimulation. We also show that affected individuals carrying this haplotype have increased serum IL-12 levels when compared with non-carriers, whereas serum IL-23 levels are decreased. We show that levels of IL-12 and IL-23 protein are increased in psoriatic lesions and that IL12B haplotype carriers have an increased IFN-γ signature in lesional psoriatic skin. These findings suggest that the cytokine environment in psoriatic lesions directs an increase in p40 expression associated with the IL12B risk haplotype, toward IL-12 at the cost of IL-23, resulting in amplification of the IFN-γ environment in lesional skin. These data emphasize the pathologic role of the Th1 axis in psoriatic pathogenesis and may have implications in terms of future targeted therapeutics.
RESULTS
The IL12B risk haplotype influences IL-12 and IL-23 serum levels
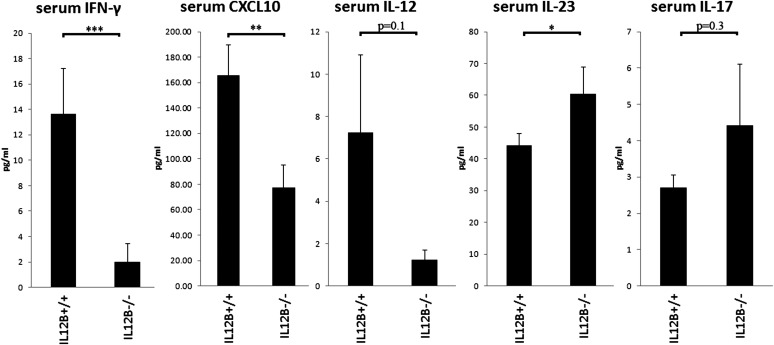
To determine if the risk-associated haplotype affects serum levels of IL-12 and IL-23 in psoriatic patients, we first addressed whether serum levels of IL-12p70, IL-22 and IL-23 differed between cases and controls. Both IL-12 and IL-23 are heterodimeric cytokines, with IL-12 (IL-12p70) being composed of the p35 and p40 subunits encoded by the genes IL12B and IL12A, respectively, whereas IL-23 is composed of the p19 (encoded by IL23A) and the p40 subunits. IL-12 levels were higher in affected cases when compared with controls, although this did not reach significance. No difference was observed with IL-23 serum levels between cases and controls, whereas IL-22 levels were significantly higher in affected patients (P < 0.05) (Supplementary Material, Fig. S1). To address the effect of the psoriasis-associated IL12B risk haplotype, as defined by the G allele of rs6887695 and the A allele of rs32112227A (15), on the serum levels of these two cytokines, we compared the serum cytokine levels in affected homozygous risk haplotype carriers against the levels in affected non-carriers. We found IL-12p70 serum levels to be higher in homozygous carriers when compared with non-carriers (6-fold, P = 0.10), whereas IL-23 levels were decreased (1.4-fold, P < 0.05). We then compared serum levels of IFN-γ and the IFN- γ induced chemokine; CXCL10 between the two groups, finding that IFN-γ was about 6.9-fold higher in homozygous risk carriers when compared with non-carriers (P < 0.0001), and CXCL10 was 2.1-fold higher (P < 0.01). Consistent with serum IL-23, IL-17A levels were lower in homozygous risk carriers when compared with non-carriers (1.6-fold), although this did not reach statistical significance. IL-22 levels did not differ between the two groups (Supplementary Material, Fig. S2). (Fig. 1)
Figure 1.
Increased levels of IFN-γ- and the IFN-γ-induced chemokine; CXCL10 were observed in serum of affected IL12B risk haplotype carriers (rs6887695G/ rs3212227A) when compared with non-carriers. Similarly, IL-12 levels were increased about 6-fold, whereas IL-23 levels were decreased about 1.4-fold. Similar to IL-23, serum IL-17 levels were down about 1.6-fold, although this was not significant (n = 202 affected homozygous risk carriers, n = 17 affected non-carriers). All values are expressed as mean ± SEM. Statistical significance of two-tailed t-test is indicated by one (P < 0.05), two (P < 0.01) or three (P < 0.001) asterisks.
IFN-γ pre-stimulation enhances IL-12 and IL-23 mRNA expression and secretion
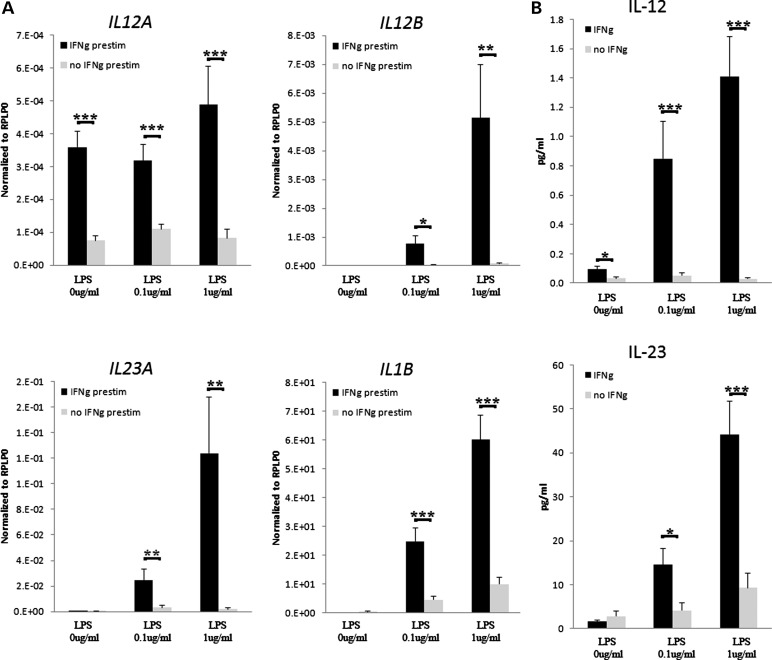
Monocytes and dendritic cells are the primary source of IL-12 and IL-23 (16). To better reproduce the conditions in psoriatic skin, we compared IFN-γ pre-stimulated and non-stimulated monocytes that were stimulated with lipopolysaccharide (LPS) at 0.1 and 1.0 μg/ml for 24 h. Consistent with our previously published findings (17), IFN-γ stimulation promotes increased expression of IL12B, IL12A, IL23A and IL1B mRNAs from monocytes in a stimulation-dependent manner (Fig. 2). The fold increase in expression was highest for IL23A at the 12 h time point (140-fold, P < 0.01, Supplementary Material, Fig. S5), whereas IL12B was more strongly induced at 24 h (98-fold, P < 0.01) (Fig. 2A). Similarly, IFN-γ stimulation increased LPS-induced secretion of both IL-12 and IL-23 proteins (P < 0.001, Fig. 2B), although the fold change was more pronounced for IL-12 when compared with IL-23 (47-fold versus 5.4-fold at 24 h, 1 ug/ml LPS).
Figure 2.
Pre-stimulation of monocytes with IFN-γ (50 ng/ml) leads to increased mRNA expression of IL12B, IL12A, IL23A and IL1B after 24 h of LPS stimulation (A). Similarly, protein levels of IL-12 and IL-23 were increased at 24 h in monocyte-conditioned medium after IFN-γ pre-stimulation. Gene expression values (mRNA) are shown relative to the housekeeping gene RPLP0 as mean ± SEM (n = 20 for IL-12 and n = 35 for IL-23). Statistical significance of two-tailed t-test is indicated by one (P < 0.05), two (P < 0.01) or three (P < 0.001) asterisks.
The IL12B risk haplotype leads to increased IL12B mRNA expression in IFN-γ conditioned monocytes
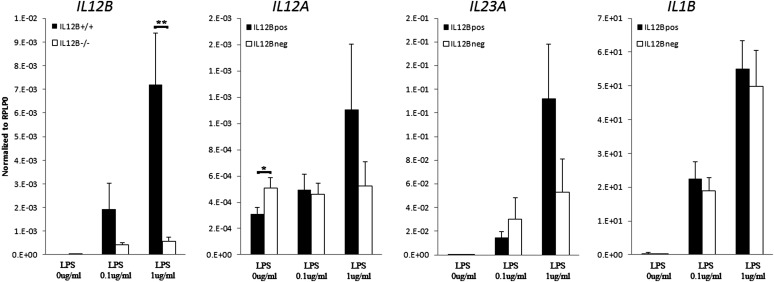
As shown in Figure 3, homozygous carriers of the IL12B risk haplotype (rs6887695 G/rs32112227 A) had consistent increases in IL12B expression in both IFN-γ primed cultures, when compared with non-risk carriers (12.5-fold, P < 0.01; 3.8-fold P < 0.01, respectively, at 24 h time point, Fig. 3). IL12A had slightly lower expression in unstimulated IFN-γ primed cultures at 24 h (1.6-fold down, P < 0.05). No significant differences were observed between risk haplotype carriers and non-carriers in terms of IL23A or IL1B mRNA expression (Fig. 3). No consistent differences were observed in the secretion of IL-12 and IL-23 proteins in culture supernatants between the two groups.
Figure 3.
Homozygous IL12B risk haplotype (rs6887695G/rs3212227A) carriers had on average 12.5-fold higher IL12B expression after 24 h stimulation when compared with homozygous non-carriers. IL12A had slightly decreased expression in unstimulated cultures. In contrast, there were no consistent changes observed in the expression of either IL23A or IL1B between the two groups. All values are expressed relative to the housekeeping gene RPLP0 as mean ± SEM (n = 29 homozygote carriers and 15 non-risk carriers). Statistical significance of two-tailed t-test is indicated by one (P < 0.05) or two 9 P < 0.01) asterisks.
IL-12 is up-regulated in psoriatic skin
To assess whether the levels of transcripts encoding the subunits of IL-12 and IL-23 are altered in psoriatic skin, we performed quantitative real time-polymerase chain reaction (QRT-PCR) on RNA isolated from healthy control, uninvolved and lesional psoriatic skin. Consistent with previous findings (16), increased expression of IL12B and IL23A mRNA was observed in lesional skin (6.7-fold and 4.9-fold, respectively, P < 0.01), whereas IL12A mRNA levels were down-regulated (3.1-fold, P < 0.05) when compared with uninvolved skin (Fig. 4). To confirm this at the protein level, skin lysates from healthy control, uninvolved and lesional psoriatic skin were obtained, and soluble IL-12 and IL-12p40 were measured by multiplex immunoassays. IL-12p70 levels were similar between control and non-lesional skin, but were approximately 4-fold higher in lesional skin (P < 0.01, Fig. 4). Likewise, protein levels of the IL-12p40 subunit were increased by about 4-fold in lesional when compared with non-lesional skin (P < 0.01) (Fig. 4). IL-12p40 was approximately five to six times more abundant than IL-12p70 (P < 0.01, Fig. 4) per milligram total protein tissue lysate.
Figure 4.
Expression of IL12B and IL23A mRNA were increased in lesional psoriatic skin, whereas IL12A expression was decreased (A) (n = 7–10). Measurements of IL-12 (IL-12p70) and IL-12p40 from tissue lysates obtained from control (NN), uninvolved (PN) and lesional psoriatic skin (PP) (n = 9–13) revealed significantly increased protein levels of both IL-12p70 and IL-12p40 in lesional psoriatic skin (B). All mRNA values are expressed relative to the housekeeping gene RPLP0 as mean ± SEM. Protein values are expressed relative to total protein tissue lysate (1 mg/ml) as mean ± SEM. Statistical significance of two-tailed t-test indicated by one (P < 0.05) or two (P < 0.01) asterisks.
Enrichment of IFN-γ induced genes in lesional skin of IL12B risk allele carriers
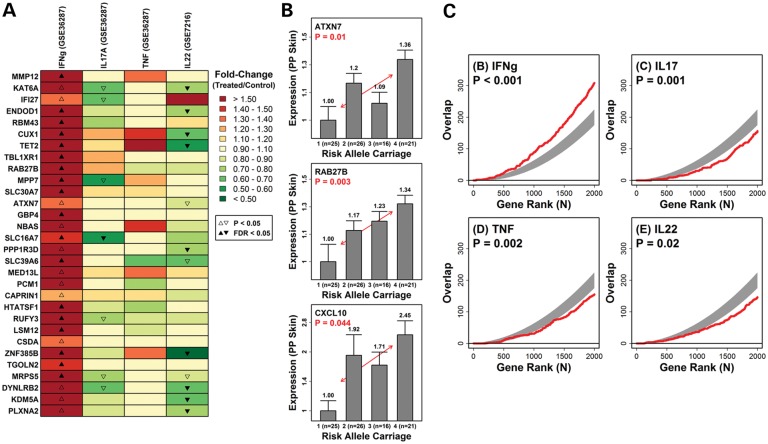
To ask whether the IL12B risk allele would lead to shifts in the cytokine environment in psoriatic skin, we assessed genome-wide expression profiles of lesional skin samples from 53 psoriasis patients (18). We identified 356 genes for which expression was significantly elevated in the lesional skin of patients in proportion to IL12B risk allele carriage (number of rs6887695 G alleles + number of rs3212227 A alleles, n = 53; P < 0.05) (Fig. 5). Of the 30 genes showing the highest fold change in lesional skin from IL12B risk allele positive individuals, several included known cytokine-responsive genes, including CXCL9 and CXCL10 (Fig. 5A and B). We, therefore, evaluated whether there was a significant tendency for genes elevated in lesional skin from IL12B risk carriers to also be induced by the cytokines IFN-γ, IL-17, TNF-α and IL-22 in cultured keratinocytes (Fig. 5C). Our analysis revealed that the top-ranked genes showing a trend toward elevated expression in lesional skin of IL12B risk carriers overlapped significantly with genes most strongly induced by IFN-γ in cultured keratinocytes (P < 0.001; Fig. 5A and C). In contrast, genes induced by IL-17 and IL-22 had significantly decreased overlap (P < 0.001; Fig. 5A and C), and a modest decrease in overlap was observed for TNF-α induced genes (P = 0.028). These analyses show that genes with elevated expression in lesional skin of IL12B risk carriers are significantly more likely to be induced by IFN-γ, but not by IL-17 or IL-22. This pattern was manifested by a number of IL12B risk-associated genes, each of which was induced by IFN-γ in cultured keratinocytes, despite weak or significant repression of the same genes by IL-17, TNF-α or IL-22 (Fig. 5A and C). These results are consistent with deviation of the inflammatory network toward IFN-γ and Th1 responses at the cost of IL-17 and IL-22 in IL12B risk allele carriers (Supplementary Material, Fig. S3 and S4).
Figure 5.
We identified 356 genes for which expression was significantly elevated in the lesional skin of patients in proportion to IL12B risk allele carriage (0–4 risk alleles, including rs6887695 G allele and rs3212227 A allele; n = 53 patients; P < 0.05). The heat map shows 30 genes with increased expression in lesional skin from individuals with more IL12B risk alleles (rs6887695 and rs3212227, P < 0.035) and the expression responses of these genes to IFN-γ, IL-17, TNF-α and IL-22 in cultured keratinocytes (A). Expression levels of three representative genes demonstrate a dose-dependent increase in the expression levels in psoriatic skin based on risk allele carriage. The red line denotes the least squares regression estimate with a positive slope in each case. (B) Furthermore, all genes were ranked according to their level of positive association with IL12B risk carriage (rs6887695 and rs3212227), and this list was compared for overlap with gene lists ranked by their level of induction by IFN-γ, IL-17, TNF-α or IL-22 in keratinocytes (C). In each box, the red line shows the level of overlap among the top n genes taken from each list, whereas the gray region outlines the central 95% of the null distribution under a random sampling model (hypergeometric distribution). Larger than expected overlap (for given n) is present, if the red line lies above the gray region, whereas lower than expected overlap (for given n) is present, if the red line lies below the gray region. P-values are based upon the overlap between the top 1000 genes from each gene list.
DISCUSSION
The involvement of IFN-γ in psoriasis pathogenesis is widely appreciated (19–21), and for a long time, psoriasis was believed to be primarily a Th1, IFN-γ-mediated disease based on the significant IFN-γ expression in psoriatic skin (20) and Th1 dominance of circulating and lesional T-cells (22). The key cytokine that polarizes T-cells toward the Th1 phenotype is IL-12 (8)that is composed of the p40 and p35 protein subunits encoded by the IL12B and IL12A genes, respectively. Both chains of the IL-12 receptor, IL-12Rβ1 and IL-12Rβ2, are up-regulated in psoriasis (23), suggesting that this cytokine may have an active role in the disease pathogenesis. However, as IL12A mRNA expression is generally down-regulated in psoriatic lesions (16) (Fig. 4A), whereas IL12B and IL23A mRNAs are increased, this has been interpreted as IL-12 playing a more limited role in psoriasis when compared with IL-23 (16).
The role of IL-23 appears to be primarily through its effect on Th17 responses. In contrast to the direct effect of IL-12 on Th1 differentiation (24), IL-23 does not act directly on naïve T cells to induce Th17 differentiation (25,26), but instead up-regulates IL-17 production and promotes survival and expansion of activated Th17 cells. These cells have been shown to be highly pathogenic and essential for the establishment of autoimmune inflammation in mouse models (27). Likewise, effective management of psoriasis is linked to suppression of IL-17 signaling (28,29), and genetic findings from genome-wide association studies have implicated genes that are more strongly involved in IL-23/IL-17 signaling than those for IL-12 (5,30). Likewise, in a xenograft system, where uninvolved skin spontaneously transforms into psoriatic lesions, blockade of IL-23 prevents the development of psoriasis (31).
Taken together, these findings have resulted in the focus being shifted toward IL-17 and a Th17-centric model of psoriasis (32). However, the relationship between IFN-γ and IL-17 is complex. Th1 and Th17 cells are often co-localized in pathologic environments (33,34), and Th1 cells are reported to inhibit Th17 development through IFN-γ (26,35,36). Moreover, IFN-γ can act on resident dendritic cells to promote the induction and expansion of Th17 cells as well as their recruitment, through expression and production of CCL20 (17). In addition, Th17 cells demonstrate considerable plasticity, and IFN-γ and IL-12 have been shown to synergize to convert Th17 cells into Th17-Th1 cells (33,37). These Th17-Th1 cells may have a more important pathologic role as recent data suggest that Th17 cells become pathogenic only after switching to a Th1 phenotype (38,39).
Both IFN-γ- and IL-17-secreting cells are found in lesional psoriatic skin, with IFN-γ positive cells being about nine times more frequent than those producing IL-17(34). Psoriatic dendritic cells are able to induce a population of activated T cells that simultaneously produce IL-17 and IFN-γ, which is not seen with dendritic cells from normal skin(40), suggesting that psoriasis is a mixed Th1 and Th17 inflammatory disease. Interestingly, a single intradermal injection of IFN-γ can promote development of an inflammatory environment that, in some respects, parallels that observed in psoriatic lesions (41). Likewise, IFN-γ expression has been shown to be increased in uninvolved skin (17,41,42), and becomes increasingly prominent as psoriasis becomes more severe (41). This suggests that psoriasis patients have greater propensity to produce IFN-γ when compared with healthy controls (41) and that IFN-γ may have a crucial role in psoriatic pathogenesis (41).
Our data are consistent with a model in which increased expression of p40 (IL12B) leads to increased production of IL-12, resulting in increased serum levels of IFN-γ and amplification of the IFN-γ signature in lesional psoriatic skin (Fig. 6). No clinical trials are available on the effect of neutralization of IFN-γ in psoriasis, but the effect of treatments neutralizing IL-17 is unmistakable and associated with a very strong clinical response with a good proportion of patients achieving clinical remission while on treatment (43,44). Although our data cannot determine whether the pathogenic effect of the IL12B risk haplotype is directly mediated by Th1 cells, or through accelerated survival and expansion of Th17 cells to a more pathogenic Th1 phenotype, it is tempting to speculate that the role of the IL12B risk allele is to prime the skin for induction of the psoriatic process through increased expression of IFN-γ that may also amplify the inflammatory response in concert with IL-17. Taken together, our data provide insight into how genetic variation at the IL12B locus influences the inflammatory network in psoriasis, reemphasizing the importance of the Th1 axis with implications for the development of targeted therapeutics.
Figure 6.

Proposed model of the pathologic role of the IL12B risk locus in psoriasis. Under normal circumstances, there is a balance between IL-12 and IL-23 production by monocytes. When monocytes/dendritic cells are exposed to IFN-γ, there is a shift toward increased IL-12 production, although there is concomitant, but less, increase in IL-23 production. In psoriatic lesions, which are rich in IFN-γ, the increase in the expression of IL12B mRNA as a function of the IL12B risk haplotype is shifted toward IL-12, leading to an even greater increase in IFN-γ that locks the mechanism in a positive feedback cycle favoring IL-12 and resulting in progressive increase in the strength of the IFN-γ signature in inflamed psoriatic skin.
MATERIALS AND METHODS
Study population
Forty-nine individuals were identified and recruited from our extended genetic cohort, and these were evenly distributed between cases and controls (25 healthy individuals versus 24 psoriatics and between risk allele carriers versus non-carriers). Thirty-three were homozygous carriers and 16 were non-carriers of the risk haplotype in the IL12B gene (rs6887695 G allele and rs3212227 A allele). Typing was performed as described in Nair et al. (15). In addition, 11 healthy controls and 13 psoriatic patients were recruited for skin biopsies (6 mm) of normal, uninvolved and lesional psoriatic skin. Informed consent was obtained from all subjects, under protocols approved by the Institutional Review Board of the University of Michigan. This study was conducted in compliance with good clinical practice and according to the Declaration of Helsinki Principle.
Microarrays
The subjects enrolled for gene expression analysis have been previously described in accord with a protocol approved by the Institutional Review Board of the University of Michigan. Shortly, it involved 58 subjects with untreated chronic plaque psoriasis with all samples run on HU133 Plus 2.0 arrays (Affymetrix, Foster City, CA, USA) as described (45). Patients were typed for risk allele polymorphisms in the IL12B gene as described (15). The raw data from the psoriatic microarrays were processed using the Robust Multichip Average method (46) and adjusted to account for gender and batch effect (patient samples only). The raw and normalized expression data are available from the Gene Expression Omnibus database (accession GSE13355).
Keratinocyte cultures
Normal human keratinocyte (NHK) cultures were established from sun-protected adult human skin as described (47) from three separate donors. Keratinocytes were grown in serum-free medium optimized for high-density keratinocyte growth (Medium 154; Invitrogen/Cascade Biologics, Portland, OR, USA). NHKs were used for experiments in the second or third passage. All cells were plated at 5000 cells/cm2 and maintained to 4 day post-confluency. Cultures were then starved of growth factors in unsupplemented medium M154 for 24 h before cytokine stimulation. NHKs were stimulated with TNF-α (10 ng/ml), IFN-γ (50 ng/ml) and IL-17(10 ng/ml) (R&D Systems, Minneapolis, MN, USA) or unstimulated control. Experiments were carried out under low-calcium (0.1 mm) conditions. Cells were harvested after 24 h, and RNA was isolated and extracted using RNeasy columns (Qiagen, Chatsworth, CA). RNA quality was checked using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) before running on Affymetrix HU133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA, USA).
Monocyte cultures
Monocytes were isolated from peripheral blood mononuclear cells by negative selection beads (Miltenyi Biotec, Auburn, CA, USA). Purity of cells was evaluated and found to be >75%. Monocytes were cultured for 72 h in Roswell Park Memorial Institute containing 10% fetal calf serum supplemented with IL-6 (5 ng/ml) (R&D Systems), IL-10 (5 ng/ml) (R&D Systems) and M-CSF (10 ng/ml) (R&D Systems) with or without IFN-γ (50 ng/ml) (R&D Systems). The monocytes were then stimulated with varying doses of LPS (0, 0.1 and 1 ng/ml) for 0, 12 and 24 h. RNA and supernatants were harvested at those time points for ELISA and multiplex assays for IL-12 (IL12p35, eBioscience, San Diego, CA, USA), IL-23 (IL23p19, MilliplexTM Map Kit Human Cytokine Panel, Millipore) and IL-1B (Fluorokine MAP human IL-1β, R&D Systems).
Tissue processing
Normal and uninvolved skin biopsies (6 mm) were obtained from sun-protected skin of healthy individuals, whereas lesional skin was obtained from active psoriatic lesions. Skin was anesthetized with lidocaine with epinephrine (1:10 000), and biopsies were snap frozen in liquid nitrogen and stored at −80°C until processing. For protein quantification, biopsies were pulverized and dissolved in complete radioimmunoprecipitation buffer for protein quantitation. Tissue lysates for protein quantitation were normalized to 1 mg/ml of total protein before analysis. For total RNA extraction, biopsies were pulverized, and RNA extraction was performed using RNAeasy kit (Quiagen) using glass beads (Biospec Products, Bartlesville, OK, USA) for homogenization. RNA quantity and quality were measured as described below.
QRT-PCRs
RNA was isolated from stimulated monocytes and extracted using RNeasy columns (Qiagen). RNA quantity and quality were measured on an Agilent 2100 Bioanalyzer (Agilent Technologies), and only samples yielding intact 18S and 28S ribosomal RNA profiles were used. Reversed transcription was performed using High Capacity cDNA Transcription kit (Applied Biosystems Inc., Foster City, CA, USA) as previously described. Transcripts were quantified using a 7990HT Fast Real-Time PCR system (Applied Biosystems) using Taqman primers sets purchased from Applied Biosystems (IL12A Hs00168408_m1, IL12B Hs01011510_m1, IL23A Hs00372324_m1, RPLP0 Hs99999902_m1 and IL1B Hs00174097_m1). All values were normalized to the expression of the housekeeping gene ribosomal protein, large, P0 (RPLP0).
Serum ELISA
Frozen serum was obtained from archived samples from 227 affected and 55 healthy individuals. ELISA was performed for IL-12 (IL12p35, eBioscience, San Diego, CA, USA), IL-22 (IL-22 DuoSet, R&D Systems), CXCL10 (CXCL10 DuoSet, R&D Systems), IL-23 (IL23p19, MilliplexTM Map Kit Human Cytokine Panel, Millipore, Billerica, MA, USA), IL-17 (IL-17, eBioscience) and IFN-γ (Quantikine, R&D Systems).
Statistical analysis
Data were tested for normality using the Kolmogorov–Smirnov test, and statistical significance was calculated using Student's t-test or Mann–Whitney test as appropriate using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). Genes associated with IL12B risk allele carriage in lesional skin were identified using least squares regression. For each probe set, log2-transformed expression was treated as a continuous response variable, and total risk allele carriage was treated as the predictor variable (n = 53 patients). Total risk allele carriage was calculated by pooling the number of risk alleles across two IL12B SNPs (i.e. number of rs6887695 g alleles + number of rs3212227 A alleles). Among the 53 subjects, 4 (8%) carried 1 risk allele, 12 (23%) carried 2, 16 (30%) carried 3 and 21 (40%) carried 4. Overlap between genes positively associated with IL12B risk allele carriage and genes induced by cytokines in vitro was assessed by the comparison of ranked gene lists (48). For each analysis, we considered only genes positively associated with IL12B risk allele carriage (slope >0) and ranked these genes according to the P-value obtained from the least squares regression analysis. This list was evaluated for overlap with genes ordered by their level of induction by IFN-γ, IL-17, TNF or IL-22 in cultured keratinocytes (Gene Expression Omnibus accessions GSE7216 and GSE36287). For each cytokine, only induced genes were considered (treated/control fold-change >1), and genes were ranked according to P-values. These P-values were generated from two-sample comparisons (i.e. treated versus control) and Bayesian linear modeling methods, as implemented in the Limma package developed for the R statistical software package (49).
SUPPLEMENTARY MATERIAL
FUNDING
J.E.G. was funded in part by grants from the American Skin Association, National Psoriasis Foundation, Dermatology Foundation, A. Alfred Taubman Medical Research Institute by the Frances and Kenneth Eisenberg Emerging Scholar Award and the National Institute of Health (K08AR060802). A.J. was supported by an American Skin Association Research Scholar Award. J.T.E. and R.P.N. and P.E.S. acknowledge support from the National Institute of Health (R01 AR04742, AR050511 and AR054966). J.T.E. was supported by the Ann Arbor Veterans Affairs Hospital. This work was also supported by the Babcock Endowment Fund at the University Of Michigan Department Of Dermatology.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Anna Pero and Sarah LaPonsa for excellent subject recruitment.
Conflict of Interest statement. The authors have no conflict of interest to the data presented in this manuscript.
REFERENCES
- 1.Gudjonsson J.E., Elder J.T. Psoriasis: epidemiology. Clin. Dermatol. 2007;25:535–546. doi: 10.1016/j.clindermatol.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Nestle F.O., Kaplan D.H., Barker J. Psoriasis. N. Engl. J. Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 3.Elder J.T., Bruce A.T., Gudjonsson J.E., Johnston A., Stuart P.E., Tejasvi T., Voorhees J.J., Abecasis G.R., Nair R.P. Molecular dissection of psoriasis: integrating genetics and biology. J. Invest. Dermatol. 2010;130:1213–1226. doi: 10.1038/jid.2009.319. [DOI] [PubMed] [Google Scholar]
- 4.Capon F., Burden A.D., Trembath R.C., Barker J.N. Psoriasis and other complex trait dermatoses: from Loci to functional pathways. J. Invest. Dermatol. 2012;132:915–922. doi: 10.1038/jid.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cargill M., Schrodi S.J., Chang M., Garcia V.E., Brandon R., Callis K.P., Matsunami N., Ardlie K.G., Civello D., Catanese J.J., et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubler U., Chua A.O., Schoenhaut D.S., Dwyer C.M., McComas W., Motyka R., Nabavi N., Wolitzky A.G., Quinn P.M., Familletti P.C., et al. Coexpression of two distinct genes is required to generate secreted bioactive cytotoxic lymphocyte maturation factor. Proc. Natl. Acad. Sci. USA. 1991;88:4143–4147. doi: 10.1073/pnas.88.10.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oppmann B., Lesley R., Blom B., Timans J.C., Xu Y., Hunte B., Vega F., Yu N., Wang J., Singh K., et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 8.Boniface K., Blom B., Liu Y.J., de Waal Malefyt R. From interleukin-23 to T-helper 17 cells: human T-helper cell differentiation revisited. Immunol. Rev. 2008;226:132–146. doi: 10.1111/j.1600-065X.2008.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonardi C.L., Kimball A.B., Papp K.A., Yeilding N., Guzzo C., Wang Y., Li S., Dooley L.T., Gordon K.B. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1) Lancet. 2008;371:1665–1674. doi: 10.1016/S0140-6736(08)60725-4. [DOI] [PubMed] [Google Scholar]
- 10.Papp K.A., Langley R.G., Lebwohl M., Krueger G.G., Szapary P., Yeilding N., Guzzo C., Hsu M.C., Wang Y., Li S., et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 52-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 2) Lancet. 2008;371:1675–1684. doi: 10.1016/S0140-6736(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 11.Morahan G., Huang D., Ymer S.I., Cancilla M.R., Stephen K., Dabadghao P., Werther G., Tait B.D., Harrison L.C., Colman P.G. Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat. Genet. 2001;27:218–221. doi: 10.1038/84872. [DOI] [PubMed] [Google Scholar]
- 12.Stanilova S., Miteva L. Taq-I polymorphism in 3'UTR of the IL-12B and association with IL-12p40 production from human PBMC. Genes Immun. 2005;6:364–366. doi: 10.1038/sj.gene.6364213. [DOI] [PubMed] [Google Scholar]
- 13.Seegers D., Zwiers A., Strober W., Pena A.S., Bouma G. A TaqI polymorphism in the 3'UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes Immun. 2002;3:419–423. doi: 10.1038/sj.gene.6363919. [DOI] [PubMed] [Google Scholar]
- 14.Yilmaz V., Yentur S.P., Saruhan-Direskeneli G. IL-12 and IL-10 polymorphisms and their effects on cytokine production. Cytokine. 2005;30:188–194. doi: 10.1016/j.cyto.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Nair R.P., Ruether A., Stuart P.E., Jenisch S., Tejasvi T., Hiremagalore R., Schreiber S., Kabelitz D., Lim H.W., Voorhees J.J., et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J. Invest. Dermatol. 2008;128:1653–1661. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E., Trepicchio W.L., Oestreicher J.L., Pittman D., Wang F., Chamian F., Dhodapkar M., Krueger J.G. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J. Exp. Med. 2004;199:125–130. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kryczek I., Bruce A.T., Gudjonsson J.E., Johnston A., Aphale A., Vatan L., Szeliga W., Wang Y., Liu Y., Welling T.H., et al. Induction of IL-17+ T cell trafficking and development by IFN-gamma: mechanism and pathological relevance in psoriasis. J. Immunol. 2008;181:4733–4741. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudjonsson J.E., Aphale A., Grachtchouk M., Ding J., Nair R.P., Wang T., Voorhees J.J., Dlugosz A.A., Elder J.T. Lack of evidence for activation of the hedgehog pathway in psoriasis. J. Invest. Dermatol. 2009;129:635–640. doi: 10.1038/jid.2008.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjerke J.R., Livden J.K., Degre M., Matre R. Interferon in suction blister fluid from psoriatic lesions. Br. J. Dermatol. 1983;108:295–299. doi: 10.1111/j.1365-2133.1983.tb03967.x. [DOI] [PubMed] [Google Scholar]
- 20.Schlaak J.F., Buslau M., Jochum W., Hermann E., Girndt M., Gallati H., Meyer zum Buschenfelde K.H., Fleischer B. T cells involved in psoriasis vulgaris belong to the Th1 subset. J. Invest. Dermatol. 1994;102:145–149. doi: 10.1111/1523-1747.ep12371752. [DOI] [PubMed] [Google Scholar]
- 21.Livden J.K., Bjerke J.R., Degre M., Matre R. The effect of Goeckerman therapy on interferon in serum and suction blister fluid from patients with psoriasis. Br. J. Dermatol. 1986;114:217–225. doi: 10.1111/j.1365-2133.1986.tb02800.x. [DOI] [PubMed] [Google Scholar]
- 22.Austin L.M., Ozawa M., Kikuchi T., Walters I.B., Krueger J.G. The majority of epidermal T cells in Psoriasis vulgaris lesions can produce type 1 cytokines, interferon-gamma, interleukin-2, and tumor necrosis factor-alpha, defining TC1 (cytotoxic T lymphocyte) and TH1 effector populations: a type 1 differentiation bias is also measured in circulating blood T cells in psoriatic patients. J. Invest. Dermatol. 1999;113:752–759. doi: 10.1046/j.1523-1747.1999.00749.x. [DOI] [PubMed] [Google Scholar]
- 23.Trepicchio W.L., Ozawa M., Walters I.B., Kikuchi T., Gilleaudeau P., Bliss J.L., Schwertschlag U., Dorner A.J., Krueger J.G. Interleukin-11 therapy selectively downregulates type I cytokine proinflammatory pathways in psoriasis lesions. J. Clin. Invest. 1999;104:1527–1537. doi: 10.1172/JCI6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy K.M., Reiner S.L. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 25.Aggarwal S., Ghilardi N., Xie M.H., de Sauvage F.J., Gurney A.L. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 26.Bettelli E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., Kuchroo V.K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 27.Langrish C.L., Chen Y., Blumenschein W.M., Mattson J., Basham B., Sedgwick J.D., McClanahan T., Kastelein R.A., Cua D.J. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaba L.C., Suarez-Farinas M., Fuentes-Duculan J., Nograles K.E., Guttman-Yassky E., Cardinale I., Lowes M.A., Krueger J.G. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J. Allergy Clin. Immunol. 2009;124 doi: 10.1016/j.jaci.2009.08.046. 1022–1010 e1021–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaba L.C., Cardinale I., Gilleaudeau P., Sullivan-Whalen M., Suarez-Farinas M., Fuentes-Duculan J., Novitskaya I., Khatcherian A., Bluth M.J., Lowes M.A., et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J. Exp. Med. 2007;204:3183–3194. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair R.P., Duffin K.C., Helms C., Ding J., Stuart P.E., Goldgar D., Gudjonsson J.E., Li Y., Tejasvi T., Feng B.J., et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonel G., Conrad C., Laggner U., Di Meglio P., Grys K., McClanahan T.K., Blumenschein W.M., Qin J.Z., Xin H., Oldham E., et al. Cutting edge: a critical functional role for IL-23 in psoriasis. J. Immunol. 2010;185:5688–5691. doi: 10.4049/jimmunol.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Cesare A., Di Meglio P., Nestle F.O. The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J. Invest. Dermatol. 2009;129:1339–1350. doi: 10.1038/jid.2009.59. [DOI] [PubMed] [Google Scholar]
- 33.Annunziato F., Cosmi L., Santarlasci V., Maggi L., Liotta F., Mazzinghi B., Parente E., Fili L., Ferri S., Frosali F., et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowes M.A., Kikuchi T., Fuentes-Duculan J., Cardinale I., Zaba L.C., Haider A.S., Bowman E.P., Krueger J.G. Psoriasis vulgaris lesions contain discrete populations of Th1 and Th17 T cells. J. Invest. Dermatol. 2008;128:1207–1211. doi: 10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- 35.Veldhoen M., Hocking R.J., Atkins C.J., Locksley R.M., Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Mangan P.R., Harrington L.E., O'Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., Weaver C.T. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 37.Lexberg M.H., Taubner A., Albrecht I., Lepenies I., Richter A., Kamradt T., Radbruch A., Chang H.D. IFN-gamma and IL-12 synergize to convert in vivo generated Th17 into Th1/Th17 cells. Eur. J. Immunol. 2010;40:3017–3027. doi: 10.1002/eji.201040539. [DOI] [PubMed] [Google Scholar]
- 38.Bending D., De la Pena H., Veldhoen M., Phillips J.M., Uyttenhove C., Stockinger B., Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Annunziato F., Romagnani S. The transient nature of the Th17 phenotype. Eur. J. Immunol. 2010;40:3312–3316. doi: 10.1002/eji.201041145. [DOI] [PubMed] [Google Scholar]
- 40.Zaba L.C., Fuentes-Duculan J., Eungdamrong N.J., Abello M.V., Novitskaya I., Pierson K.C., Gonzalez J., Krueger J.G., Lowes M.A. Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell-polarizing myeloid dendritic cells. J. Invest. Dermatol. 2009;129:79–88. doi: 10.1038/jid.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson-Huang L.M., Suarez-Farinas M., Pierson K.C., Fuentes-Duculan J., Cueto I., Lentini T., Sullivan-Whalen M., Gilleaudeau P., Krueger J.G., Haider A.S., et al. A single intradermal injection of IFN-gamma induces an inflammatory state in both non-lesional psoriatic and healthy skin. J. Invest. Dermatol. 2012;132:1177–1187. doi: 10.1038/jid.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uyemura K., Yamamura M., Fivenson D.F., Modlin R.L., Nickoloff B.J. The cytokine network in lesional and lesion-free psoriatic skin is characterized by a T-helper type 1 cell-mediated response. J. Invest. Dermatol. 1993;101:701–705. doi: 10.1111/1523-1747.ep12371679. [DOI] [PubMed] [Google Scholar]
- 43.Leonardi C., Matheson R., Zachariae C., Cameron G., Li L., Edson-Heredia E., Braun D., Banerjee S. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N. Engl. J. Med. 2012;366:1190–1199. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 44.Papp K.A., Leonardi C., Menter A., Ortonne J.P., Krueger J.G., Kricorian G., Aras G., Li J., Russell C.B., Thompson E.H., et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N. Engl. J. Med. 2012;366:1181–1189. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 45.Gudjonsson J.E., Ding J., Johnston A., Tejasvi T., Guzman A.M., Nair R.P., Voorhees J.J., Abecasis G.R., Elder J.T. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J. Invest. Dermatol. 2010;130:1829–1840. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 47.Elder J.T., Fisher G.J., Zhang Q.Y., Eisen D., Krust A., Kastner P., Chambon P., Voorhees J.J. Retinoic acid receptor gene expression in human skin. J. Invest. Dermatol. 1991;96:425–433. doi: 10.1111/1523-1747.ep12469889. [DOI] [PubMed] [Google Scholar]
- 48.Lottaz C., Yang X., Scheid S., Spang R. OrderedList–a bioconductor package for detecting similarity in ordered gene lists. Bioinformatics. 2006;22:2315–2316. doi: 10.1093/bioinformatics/btl385. [DOI] [PubMed] [Google Scholar]
- 49.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.