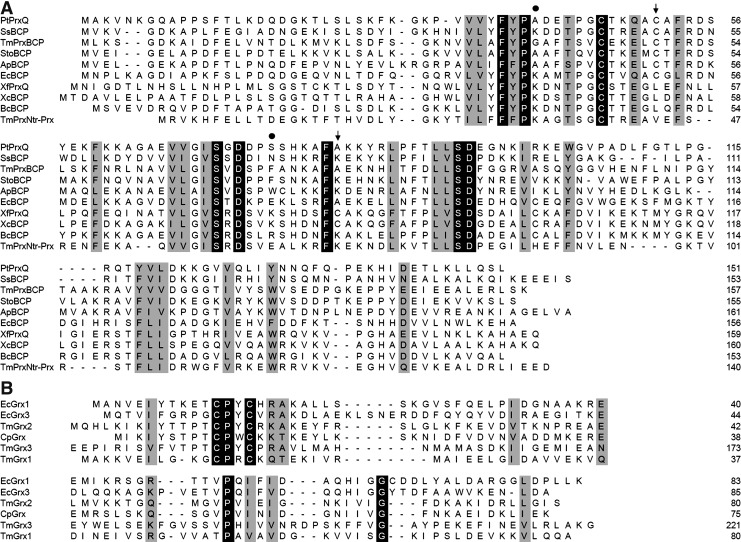
FIG. 1.
Amino acid sequence alignments of BCP/Q-type Prxs and Grx-like proteins. The alignments were done with ClustalW. In white on black: strictly conserved amino acids. In black on gray: functional amino acid conservation. (A) Sequence comparison of biochemically or structurally characterized BCPs. Accession numbers are as follows: Populus trichocarpa: PtPrxQ POPTR_0006s13980, Bulkholderia cenocepacia: BcBCP YP_002231064, Escherichia coli: EcBCP AAB88562, Xylella fastidiosa: XfPrxQ AAF83771, Sulfolobus solfataricus: SsBCP1 NP_343463, Aeropyrum pernix: ApBCP NP_148402, TmPrxNtr-Prx EHA59032, TmPrxBCP NP_228589, Xanthomonas campestris: XcBCP AEL0686, Sulfolobus tokadaii: StoBCP NP_377766. Only the BCP domain of TmPrxNtr is shown here. While the peroxidatic cysteine is strictly conserved, the position of the resolving cysteine, indicated by an arrow, can vary. In addition, some residues that are likely important for the formation of the A-type interface in dimeric BCP are indicated by a black circle. (B) Sequence comparison of Grx. Accession numbers are as follows: EcGrx3 NP_418067, EcGrx1, NP_308956, Clostridium pasteurianum CpGrx AAA23277. Only the C-terminal domain of TmGrx3 has been used for alignment. BCP, bacterioferritin comigratory protein; Grx, glutaredoxin; Ntr, nitroreductase; Prx, peroxiredoxin.