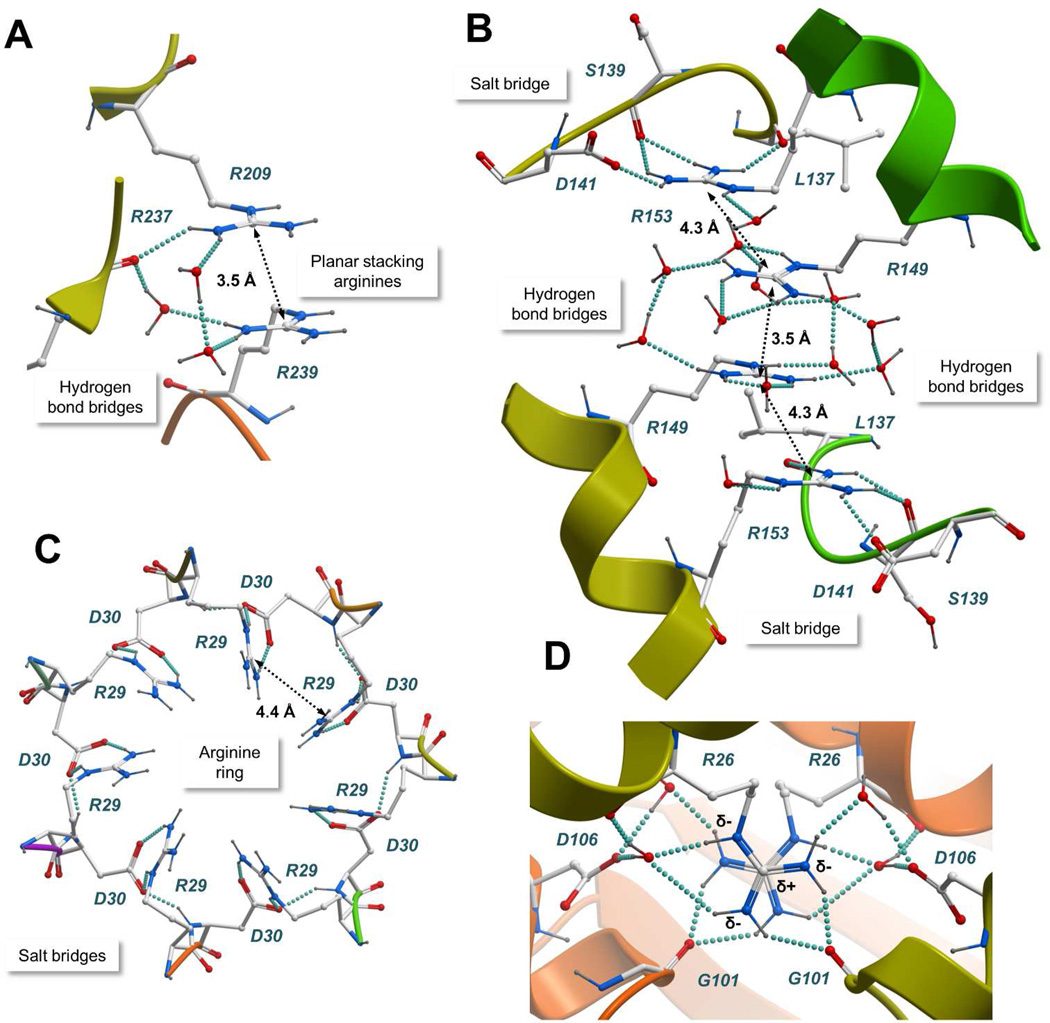
Figure 2.
Atomic details of representative arginine clusters. A) N-Acylamino acid racemase (PDB entry 1r0m); B) ATP-dependent hexokinase (PDB entry 2e2q); C) Sm protein (PDB entry 1i8f); D) Nucleoside diphosphate kinase (PDB entry 2hur). The protein structures are represented with a ribbon model and colored by subunit. Interacting residues and water molecules are shown with a “balls and sticks” model. Hydrogen bonds are represented with cyan spheres and the Cζ-Cζ distances between arginine pairs are shown. Common stabilizing interactions such as planar stacking of arginines, counterions, and bridges of hydrogen bonds are shown.