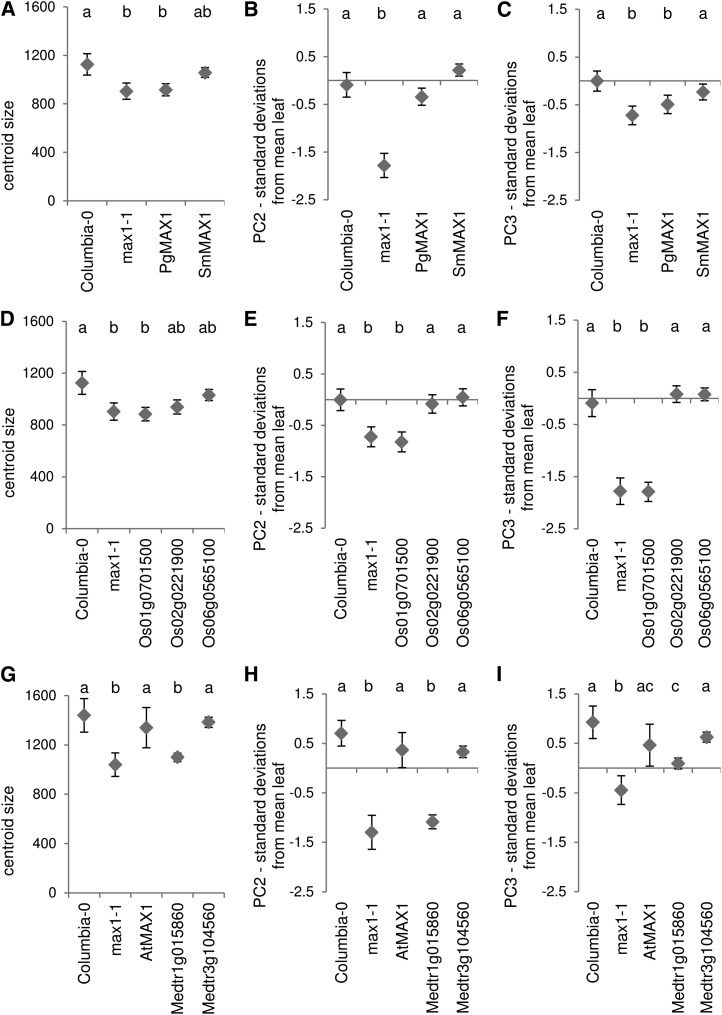
Figure 5.
Leaf phenotype rescue of Arabidopsis max1 by putative orthologs from nonangiosperm species white spruce and S. moellendorffii (A–C), in-paralogs from rice (D–F), and in paralogs from M. truncatula (G–I), compared with the Columbia wild type and max1-1. The AtMAX1 line is included as a control for the native gene function. Phenotypes used to judge rescue, as produced by LeafAnalyser, are as follows: leaf size measured by centroid size (A, D, and G) and mean number of sds of leaves from the natural accession mean for PC2 (B, E, and H) and PC3 (C, F, and I). Six to 10 plants of each of two representative lines were used per ortholog, except for the M. truncatula orthologs, in which all eight to 10 lines are included. Error bars indicate se of the mean. Shared letters indicate no significant difference in Tamhane’s T2 post hoc test (P ≤ 0.001, centroid size and PC2) or Tukey’s honestly significant difference mean-separation test (P ≤ 0.05, PC3).