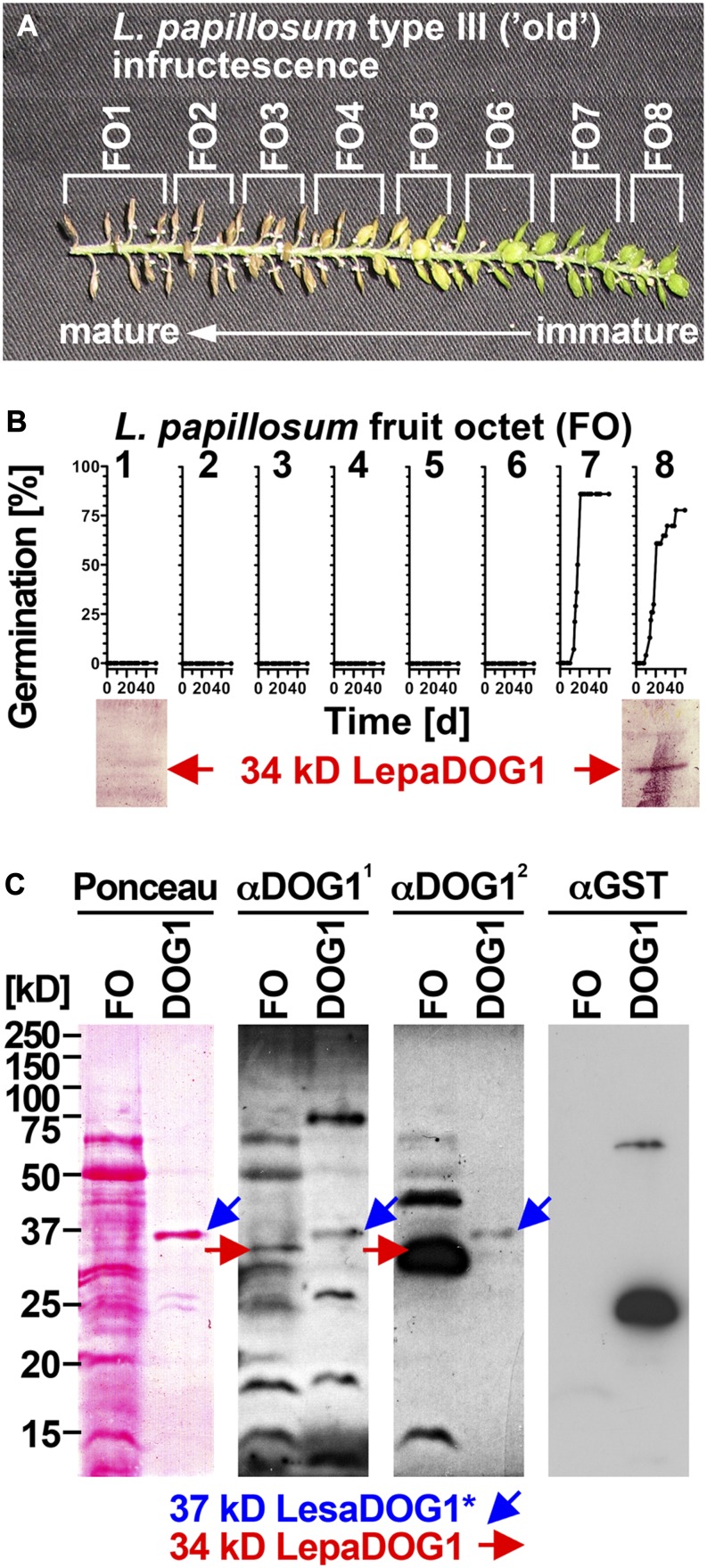
Figure 6.
Analysis of LepaDOG1 protein expression within the L. papillosum infructescence. A, Type III L. papillosum infructescence exhibiting the seed maturation gradient and the experimental dissection into FOs. B, Top panel, FO-specific germination time courses to determine the developmental switch to seed dormancy; bottom panel, FO-specific immunoblot analysis of LepaDOG1 protein (arrow) expression using a polyclonal antibody (α-DOG11) raised against AtDOG1. C, Control experiments (left panel, Ponceau-stained blotting membrane; middle and right panels, immunoblot analyses) demonstrating that two different antibodies raised against AtDOG1 (α-DOG11 and α-DOG12) detect a recombinant 37-kD LesaDOG1* antigen that corresponds to a 37-kD protein purified from an extract of E. coli expressing GST-LesaDOG1 fusion protein (DOG1; for constructs and purification procedure, see Supplemental Figs. S3 and S4). Note that in the E. coli DOG1 extract, the α-DOG1 antibodies detect the strongest 37-kD protein band (Ponceau) as LesaDOG1* (blue arrows), while this antigen is not detected by the α-GST antibody. The asterisk indicates the presence of an approximately 3-kD residual linker peptide in the recombinant LesaDOG1* protein left after GST cleavage (Supplemental Fig. S3). Therefore, an apparent molecular mass of 34 kD is expected for LesaDOG1. Both α-DOG1 antibodies detect 34-kD LepaDOG1 antigens in a L. papillosum protein extract from type I FO prior to dormancy induction (red arrows). Note that the two α-DOG1 antibodies differ in sensitivity and that the highly similar 34-kD LepaDOG1a and LepaDOG1c proteins cannot be distinguished. kD indicates the protein molecular mass marker ladder. [See online article for color version of this figure.]