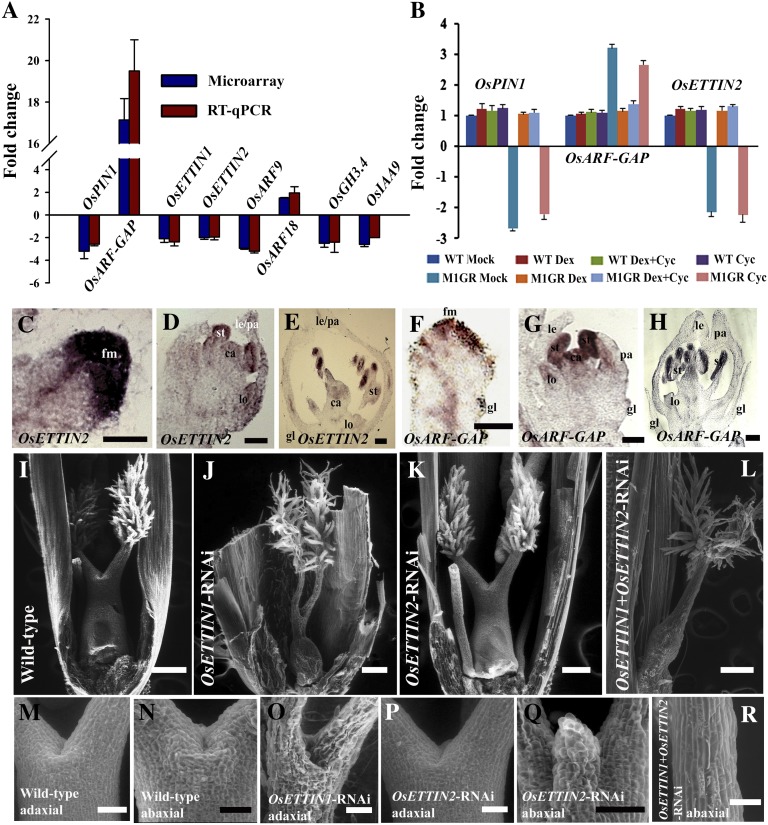
Figure 4.
Regulation of the auxin signaling pathway by OsMADS1. A, Fold change determined by RT-qPCR in the normalized expression of OsPIN1, OsARF-GAP, four ARFs (OsETTIN1, OsETTIN2, OsARF9, and OsARF18), and two auxin-responsive genes (OsGH3.4 and OsIAA9) in OsMADS1 knockdown florets. The fold change detected by microarray analysis is also shown. B, Expression levels for OsPIN1, OsARF-GAP, and OsETTIN2 in the panicles of wild-type (WT) and PUbi:OsMADS1-ΔGR; P35S:OsMADS1amiR plants treated individually with dexamethasone and cycloheximide and also in combination. The effects of these treatments were compared with that of wild-type plants. C to H, Spatial distribution of OsETTIN2 (C–E) and OsARF-GAP (F–H) transcripts in developing wild-type florets. C and F, Young FMs before organ initiation. D and G, FMs with differentiating floret organs. E and H, Florets with mature organs. Meristems and organs are labeled as in Figure 3. Bars = 50 µm in C, D, F, and G and 100 µm in E and H. I to L, Functional characterization of OsETTIN1 and OsETTIN2. Carpels are shown in the wild type (I), OsETTIN1-RNAi (J), OsETTIN2-RNAi (K), and OsETTIN1+OsETTIN2-RNAi (L). M to O, Abaxial and adaxial views of the style region above the ovary. The wild-type (M and N), OsETTIN1-RNAi (O), OsETTIN2-RNAi (P and Q), and OsETTIN1+OsETTIN2-RNAi (R) are shown. Bars = 200 µm in I to L and 50 µm in M to R.