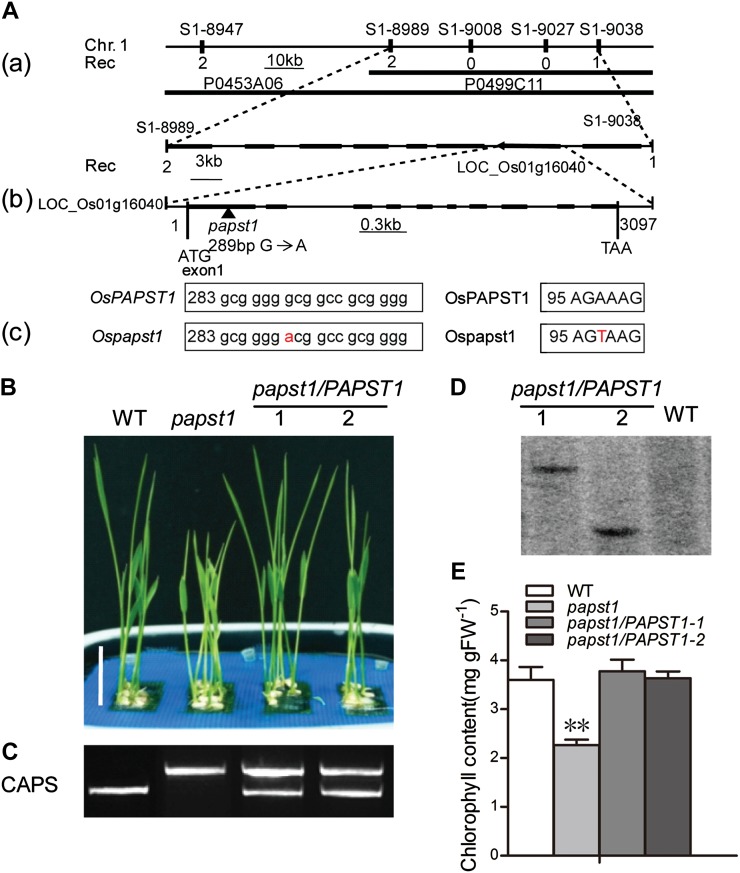
Figure 3.
Map-based cloning of OsPAPST1 and complementation test. A, Map-based cloning of OsPAPST1. OsPAPST1 was mapped between two SSR markers, S1-8989 and S1-9038, on the short arm of chromosome 1 (a). The genomic region contains eight genes (black boxes). The sequence surrounding the point mutation (G-to-A transition) in papst1 is shown in b and c. The genomic structure of OsPAPST1, comprising 10 exons and 9 introns, is indicated. B, Phenotypes of 6-d-old plants of wild-type (WT), papst1 mutant, and two transgenic plants transformed with OsPAPST1. Bar = 2 cm. C, PCR analysis using a CAPS marker for wild-type, papst1 mutant, and two transgenic plants transformed with OsPAPST1. D, Southern-blot analysis of the wild type and the two transgenic lines transformed with OsPAPST1, using the hygromycin gene as a probe. E, Chlorophyll contents of the third leaf of 6-d-old wild-type, papst1 mutant, and two transgenic plants transformed with OsPAPST1. Values represent means ± sd of 10 biological replicates. papst1 values that are significantly different from the corresponding wild-type controls are indicated by asterisks (**P < 0.01; Student’s t test). FW, Fresh weight. [See online article for color version of this figure.]