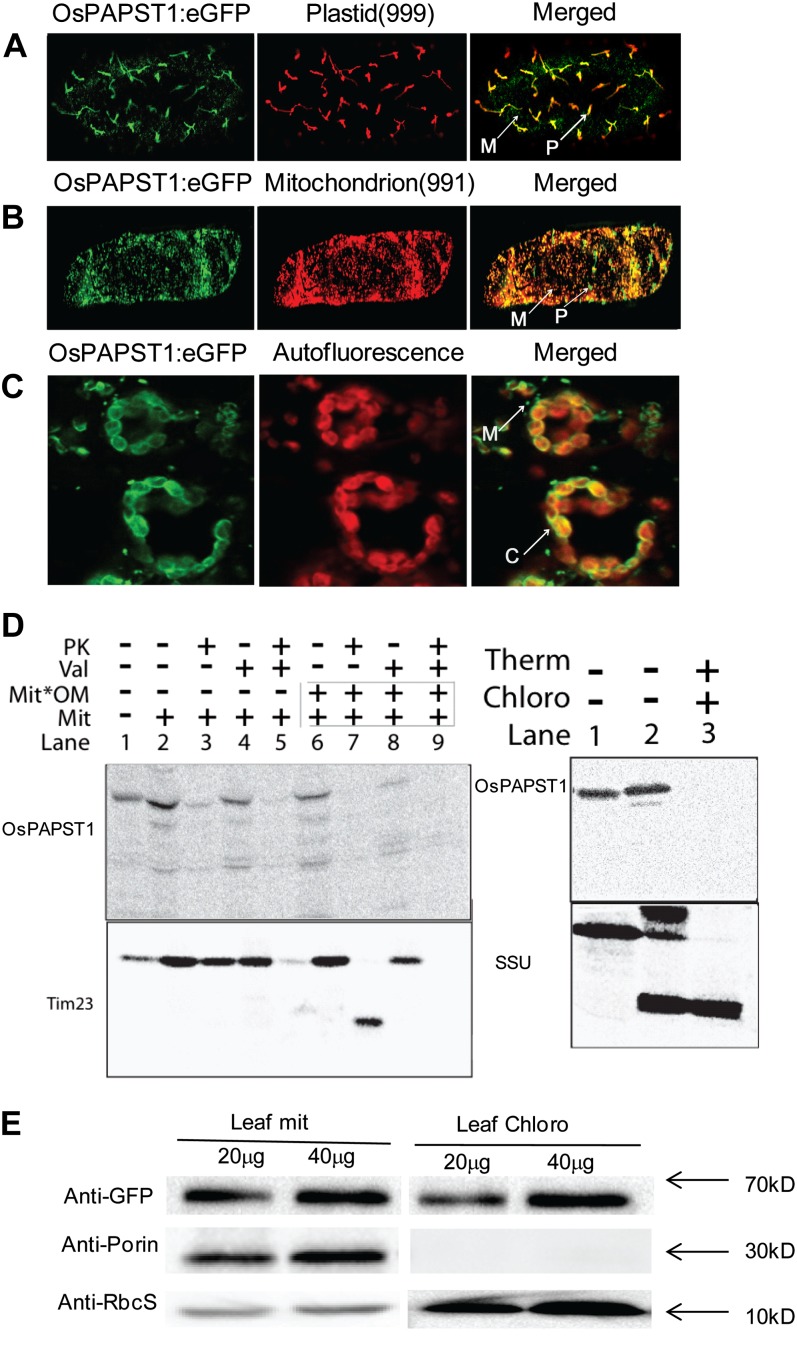
Figure 7.
Subcellular localization of OsPAPST1. A, Localization of the OsPAPST1:eGFP fusion protein in plastid indicated by a plastid marker (999; Nelson et al., 2007) in onion epidermal cells. B, Localization of the OsPAPST1:eGFP fusion protein in mitochondria indicated by a mitochondrial marker (991; Nelson et al., 2007) in onion epidermal cells. C, Crescent-shaped pattern of fluorescence of the OsPAPST1:eGFP fusion protein in chloroplast tobacco epidermal cells. P, Plastid; M, mitochondria; C, chloroplasts. D, In vitro uptake of radiolabeled OsPAPST1 into mitochondria and chloroplasts. Radiolabeled OsPAPST1 was incubated with isolated mitochondria and chloroplasts, and protease was added after 15 min where indicated. Whereas radiolabeled OsPAPST1 bound to mitochondria quite efficiently (mitochondrial panel; lane 2), it was not protected from protease digestion (mitochondrial panel; lane 3), in contrast to the inner membrane protein Tim23. A similar pattern was observed with chloroplasts, where no protease-protected protein was evident (chloroplast panel; lane 3). PK, Protease; Val, valinomycin; Mit, mitochondria; Mit*OM, outer membranes of mitochondria; Therm, thermolysin; Chloro, chloroplasts; SSU, small subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. E, Immunoblot analysis of OsPAPST1:eGFP-complemented papst1 transgenic plants. The OsPAPST1 protein was detected using an anti-GFP protein antibody. The purity of the chloroplasts and mitochondria was tested using antibodies raised against organelle-specific proteins, the small subunit of 1,5-ribulose bisphosphate for chloroplasts and porin for mitochondria.