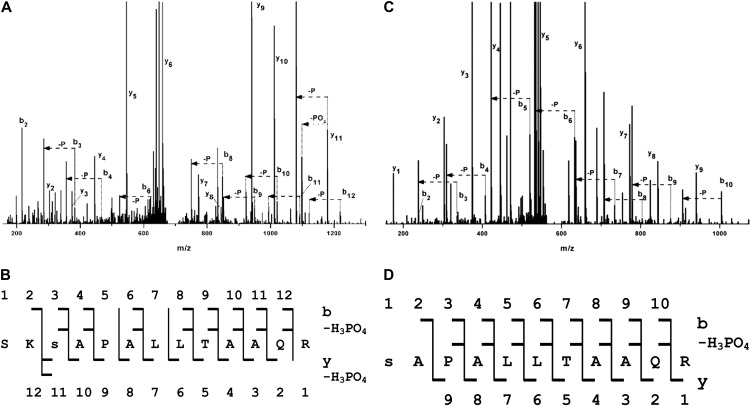
Figure 2.
The predicted 14-3-3 binding motif of HopQ1 is phosphorylated by plant kinases. A and B, HopQ1-Strep tag II fusion protein expressed in N. benthamiana leaves was affinity purified and subjected to LC-MS/MS analyses. C and D, HopQ1-6xHis expressed in bacteria was incubated with total protein extracts from N. benthamiana and then affinity purified and analyzed by LC-MS/MS. A and C show the fragmentation spectra with peak assignment to b, y, b-H3PO4, and y-H3PO4, with “–P” denoting loss of an H3PO4 group. Major signals of the MS/MS spectra are identified by their corresponding fragment tags of the b, y series, but also of the y-H3PO4 and b-H3PO4 series, which were expected in the case of a phosphorylated peptide. B and D show the peptide sequences with daughter ions of the b, y, b-H3PO4, and y-H3PO4 series found in the spectra. In the peptide derived from HopQ1 phosphorylated in vitro (C and D), the presence of the full series of b2 to b10 fragments and the expected b-H3PO4 series allows for unequivocal localization of the phosphate at Ser-51. In the peptide derived from HopQ1 expressed in planta (A and B), the majority of b and b-H3PO4 pairs are also present. In addition, the presence of strong y, y-PO3, and y-H3PO4 signals indicates that the site of phosphorylation is Ser-51, not Ser-49.