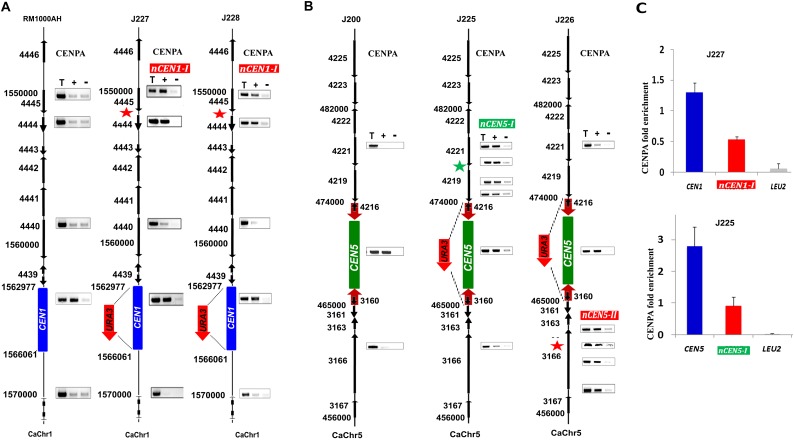
Figure 4.
Neocentromeres are preferentially formed on CEN adjacent regions on various chromosomes of C. albicans. (A) CENPA ChIP analyses in RM1000AH-cen1Δ clones mapped neocentromeres (nCENs) within 15 kb from the deleted region. (B) CENPA ChIP analyses in RM1000AH-cen5Δ clones also mapped neocentromeres (nCENs) within 15 kb from the deleted region. C. albicans carries two homologs of each chromosome. Only one homolog of Chr1 where CEN1 has been replaced by URA3 is shown. CENPA enrichment shown at the native CENs location is contributed by an unaltered homolog and is shown as a positive control for CENPA ChIP assays. Black arrows along with the Orf numbers show the gene arrangement and the direction of transcription. Red thick arrows on either side of CEN5 indicate inverted repeats. Neocentromeres on both chr1 (nCEN1-I: coordinates: Assembly 21, CaChr1 1544000–1556000) and Chr5 (nCEN5-I: coordinates: Assembly 21, CaChr5 456000–462000; nCEN5-II: coordinates: Assembly 21, CaChr5 474000–482000) formed at CEN1 proximal regions. Length of CENPA binding on Chr1 and Chr5 neocentromeres was longer (4–12 kb) than native centromeres (3–4 kb). Parent RM1000AH did not show CENPA enrichment on these neocentromere sites. (T) Total DNA; (+) with Ab; (−) without Ab (beads only). (C) CENPA ChIP qPCR analysis showing fold enrichment of CENPA on native centromeres and neocentromeres as compared with a noncentromeric region (LEU2). Enrichment of CENPA at the URA3 location was not shown here but presented as a separate figure (Supplemental Figure S5).