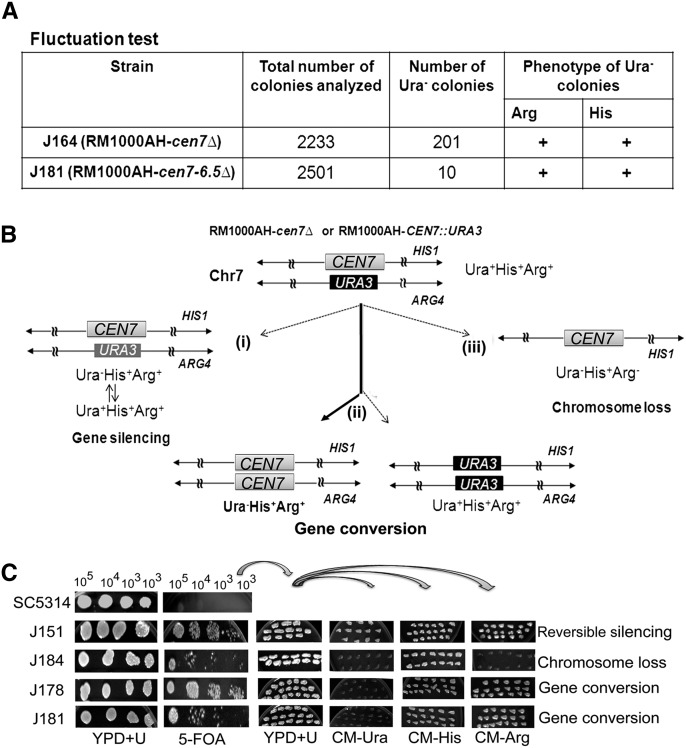
Figure 6.
Gene conversion occurs at the C. albicans centromere. (A) Fluctuation tests (see Methods) in strain J164 (RM1000AH-cen7Δ) and J181 (RM1000AH-cen7-6.5kbΔ) identified Ura−Arg+His+ colonies (bottom panel), suggesting either silencing of URA3 or gene conversion at CEN7. (B) Schematic showing possible fates of a marker on a chromosome during segregation. The Ura− phenotype can arise due to (1) silencing of URA3 placed at the centromeric context, (2) loss of the chromosome (shown here is integration of URA3 into ARG4 containing Chr7 homolog), and (3) gene conversion, which results in replacement of URA3 by CEN7 from the unaltered Chr7 or vice versa. (C) Strains SC5314 (wild-type Ura+ strain), J151 (RM1000AH-CEN7∷URA3), J184 (RM1000AH-cen7-6.5kbΔ), J164 (RM1000AH-cen7Δ), and J181 (RM1000AH-cen7-6.5kbΔ) were counter-selected on FOA plates (lower panel). SC5314 failed to grow, while J151, J184, J178, and J181 could grow on 5-FOA-containing plates. To see whether the Ura− phenotype is due to silencing, chromosome loss, or gene conversion, FOA-resistant (FOA+) colonies were revived on YPDU plates and then transferred to plates lacking uridine, histidine, or arginine. Strains J151 (RM1000AH-CEN7∷URA3) and J184 (RM1000AH-cen7-6.5kbΔ) were used as positive controls for silencing and chromosome loss, respectively. Silencing at C. albicans CEN7 is reversible (Supplemental Fig. S1B). As expected, all FOA+ colonies of J151 were Ura+Arg+His+, confirming reversible silencing. FOA+ colonies of J184 were Ura−Arg−His+, which confirms a loss of the ARG4-containing homolog of Chr 7 (shown in Table 1). FOA+ colonies of J164 and J181, however, were Ura−Arg+His+, ruling out the occurrence of silencing (Ura+Arg+His+) or chromosome loss (Ura−Arg−His+), but indicative of gene conversion. Gene conversion in Ura−Arg+His+ derivatives of both J164 and J181 was subsequently confirmed by Southern (Supplemental Fig. S7) and PCR analysis (data not shown).