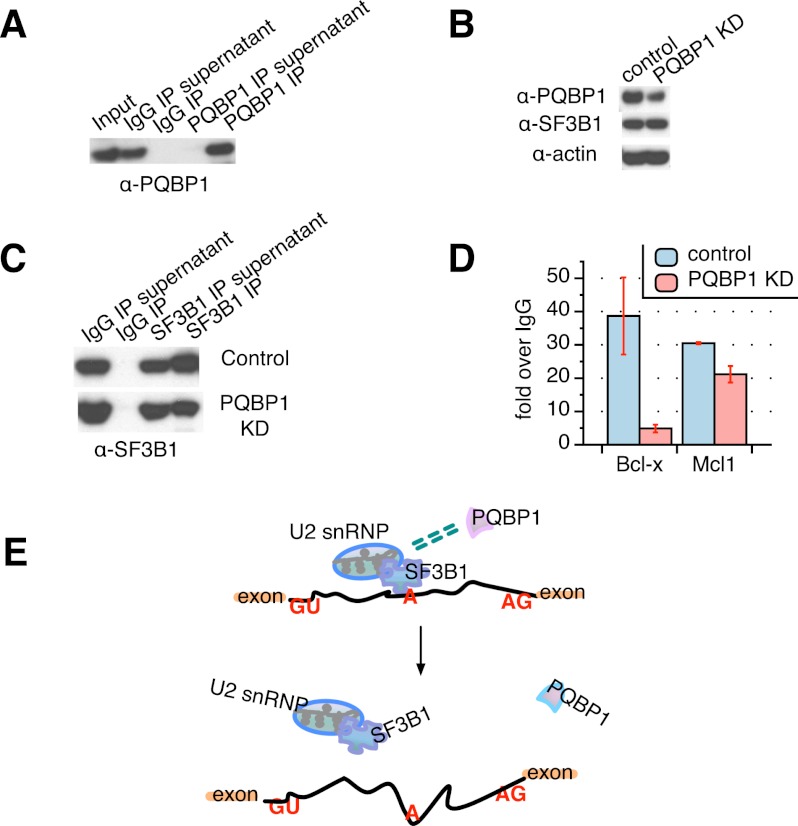
Figure 3.
SF3B1 function is affected by PQBP1. (A) Western blot of PQBP1 CLIP together with an irrelevant IgG control, probed with PQBP1 antibody. PQBP1 was not detected in depleted supernatants or the IgG pull-down, indicating efficient and specific antibody binding of PQBP1. (B) Western blot of SF3B1 CLIP from PQBP1 knockdown (KD) samples and samples treated with siRNA targeting firefly luciferase mRNA as control. Shown are total cell lysates of control (lane 1) and PQBP1 knockdown (lane 2) HeLa cells probed with antibodies indicated at the left. (C) Western blot of SF3B1 CLIP from PQBP1 knockdown samples and samples treated with siRNA targeting firefly luciferase mRNA as control. Lanes 1 and 2 are the IgG controls, and lanes 3 and 4 are SF3B1 CLIP stained by SF3B1 antibody for control and PQBP1 knockdown samples. (D) Quantification of Bcl-x and Mcl1 mRNA enrichment in the SF3B1–RNA complex as from SF3B1 CLIP in control and PQBP1 knockdown samples, respectively, by RT–PCR. Data values normalized to the IgG control and mean of three independent measurements ± SD are shown. (E) Proposed model mechanism of PQBP1 AS regulation. A schematic of part of a hypothetical mRNA is shown. Orange rectangles are exons, and the black line is the intron. “GU” marks the 5′ splice site, “A” marks the branch site recognized by the U2 snRNP with help from components like SF3B1, and “AG” marks the 3′ splice site. Green dashed double lines indicate the association between PQBP1 and SF3B1. Through an association with SF3B1 and other splicing factors, PQBP1 influences the recruitment of U2 snRNP to specific target sites, thus affecting splicing decisions on a subset of mRNAs. Loss or mutations of PQBP1 interrupt the splicing factor association and the recognition of specific splice sites. See the Discussion.