Abstract
The purpose of this study was to determine the relationship between hypermethylation of DACT1 gene promoter and lower mRNA expression in bladder urothelial carcinoma tissue. The methylation status of 29 urothelial carcinoma samples and 29 normal tissue samples were examined by methylation-specific polymerase chain reaction (MSP). The DACT1 mRNA transcript levels and DACT1 protein levels in all samples were then evaluated to define the relationship between the methylation status of the DACT1 promoter and its expression at the transcriptional and translational levels. Decreased expression of DACT1 was detected in 89.66% of urothelial carcinomas (26/29; P < 0.005). Promoter hypermethylation was found in 58.62% (17/29) urothelial carcinomas and 25% (7/29) normal tissues, respectively (P < 0.05). DACT1 expression was lower in tissues where the DACT1 gene promoter was hypermethylated than in unmethylated tissues (0.25±0.17 vs 0.69±0.30, P < 0.05). DACT1 gene hypermethylation was closely related to tumor size, grade and stage (P < 0.05). Our results indicate that silencing and downregulation of DACT1 mRNA may be implicated in carcinogenesis and the progression of bladder urothelial carcinoma, and may be a potential prognostic factor.
Keywords: DNA methylation, DACT1, hypermethylation, bladder urothelial carcinoma
INTRODUCTION
Bladder cancer is the sixth most common cancer in the world[1]. The majority of bladder cancers are comprised of urothelial carcinoma, also known as transitional cell carcinoma. Although the oncogenesis of bladder cancer is unclear so far, it is generally agreed that the accumulation of multiple genetic and epigenetic alternations leading to the activation of proto-oncogenes and/or inactivation of tumor-suppressor genes contribute to the development of bladder cancer[2]–[4].
Aberrant DNA methylation is now considered to be the most important epigenetic alteration in many cancers[5], including bladder cancer[6]. In general, DNA methylation is one of the best-studied epigenetic alterations in human cancers and may play important roles in carcinogenesis[7]. Carcinogenesis is associated with changes in this epigenetic phenomenon, including two distinct and seemingly opposing trends: global decrease in cytosine methylation (hypomethylation) and methylation of cytocine in CpG islands (hypermethylation)[8]. Such alterations can result from DNA mutations or deletions, or from epigenetic alterations, e.g., changes in gene expression that are not mediated by a change in the nucleotide sequence, such as DNA promoter hypermethylation[9]. Hypermethylation of normally unmethylated tumor suppressor genes correlates with a loss of expression in cancer cell lines and primary tumors, suggesting that hypermethylation of tumor suppressor genes could promote carcinogenesis.
The DACT1 gene was initially found as a signal regulation molecule in the clawed frog[10]. It is the principal member of the Dact family. The DACT gene is well conserved at the genomic level during evolution. The DACT1 gene maps to chromosome 14, at 14q23.1. Many studies have demonstrated that DACT1 is a key mediator in the negative regulation of the Wnt/β-catenin signaling pathway, whereas abnormal activation of this signaling pathway participates in the process of cancer progression[11]-[13]. A growing body of evidence has emerged in the past decade on the involvement of the DACT gene in the pathogenesis of cancer. Several studies have reported aberrant expression of a number of DACT genes in cancers. The examples include DACT1 in stomach neoplasms, liver cancer and germ cell tumor, and DACT2 in endometrial adenocarcinoma[14]. However, up to now, the role of DACT1 in bladder urothelial carcinoma has not yet been reported and whether DNA methylation participates in bladder carcinogenesis is also unclear.
In this study, the methylation status of the DACT1 gene was examined in tissue from 29 normal bladders and tissue from 29 bladder urothelial carcinomas. We also examined the expression level of DACT1 mRNA and protein in all samples. Using methylation-specific polymerase chain reaction (MSP) and reverse transcription polymerase chain reaction (RT-PCR), a clear relationship was found between hypermethylation status and the low expression of DACT1 in bladder transitional cell carcinomas. DACT1 was hypermethylated in almost all human bladder cancers but unmethylated in normal tissues. We hypothesized that these changes could be of diagnostic value for the detection of bladder cancer and decided to measure the degree of DNA methylation in DACT1 in bladder transitional cell carcinoma tissues.
MATERIALS AND METHODS
Tissue samples
We obtained frozen tissue samples of 29 bladder carcinomas and 29 normal bladder tissues from the First Affiliated Hospital of Nanjing Medical University. The study was approved by the Institutional Human Investigations Committee. All patients signed the informed consent. The tumors were verified as transitional cell carcinomas by two experienced pathologists according to the WHO criteria[15]. The patients had well-documented clinical history and follow-up information.
Isolation of genomic DNA
Bladder cancer tissues and normal bladder tissues were obtained after surgical resection and snap-frozen and stored at -70°C. Genomic DNA was isolated by using a Promega Wizard DNA isolation kit. The tissues were incubated at 55°C in a homogenization buffer containing 50 mmol/L Tris (pH 8.0), 1 mmol/L EDTA, 0.5% Tween 20, and 5 mg/mL proteinase K for 3 h prior to genomic DNA isolation.
RT-PCR for DACT1
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA), and cDNA was synthesized from total RNA with oligo-dT primer (Reverse Transcription System, Promega, Madison, WI, USA) following the manufacturer's instructions. Primer sequences for DACT1 amplification were 5′-GCGAAGAGATGCTGGTITGT-3′ (sense) and 5′-AAATTGTGGTGTGGAGAGGG-3′ (antisense). The PCR included 2 min of denaturation at 95°C followed by 35 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C, and a final extension for 7 min at 72°C. Primer sequences for β-actin amplification were 5′-GTGGGGCGCCCCAGGCACCA-3′ (sense); antisense: 5′-CTCCTTAATGTCACGCACGATTTC-3′ (antisense). A BLAST search confirmed that these primer sequences were identical to the endogenous human target genes. The PCR products were resolved by electrophoresis in a 1.5% agarose gel and stained with ethidium bromide.
DNA extraction and methylation-specific PCR
Genomic DNA was obtained by digestion with proteinase K (Sigma, St. Louis, MO, USA), followed by phenol/chloroform (1:1) extraction. Briefly, 4 ng genomic DNA was denatured by NaOH and modified by sodium bisulfite. The DNA was purified using a Wizard DNA purification kit (Promega), desulfonated with NaOH, precipitated with ethanol, and resuspended in water. PCR was performed with bisulfite-treated DNA as a template using specific primer sequence for methylated and unmethylated forms of the genes. The primer sequences of DACT1 for methylated reaction were as follows: 5′-ACTACTAATCAAAAACGCCCTACG-3′; (sense) and 5′-AATAGTCGTGTTTTATITTCGGGTAC-3′ (antisense). The sequences for the unmethylated reaction were 5′-AAAACTACTAATCAAAAACACCCTACAC-3′ (sense) and 5′-AT AGTTGTGTTTTATTTTTGGGTATGA-3′ (antisense).
Step-down PCR was performed in 20-µL reaction buffer containing 10×loading buffer (including Mg2+), 2 µL dNTP, 1 µL each PCR primer, 0.2 µL AmpliTaq polymerase, 2 µL bisulfite-modified DNA, and 16.8 µL diethyl-pyrocarbonate-treated water. The reaction was started by heating the samples at 95°C for 12 min followed by 35 cycles at 94°C for 30 s, 55°C (methylated) or 53°C (unmethylated) for 30 s, and 72°C for 30 s, followed by a final extension at 72°C for 7 min. The amplification products were separated on a 1.5% agarose gel and visualized by UV illumination. The results were confirmed by repeating bisulfite treatment and methylation-specific PCR for all samples.
Real-time PCR
Real-time quantitative PCR analysis was performed on cDNAs, using the ABI7300 instrument. PCR samples were prepared in a final volume of 20 µL containing TagHS (5 U/µL), 10× buffer (Mg2+ plus), dNTP mixture (2.5 µmol/L), forward primer (20 µmol/L), reverse primer (20 µmol/L), 2 µL cDNA template, and 20× Eva Green (1 µL). PCR primer sets were as follows: β-actin: sense 5′-GTGGGGCGCCCCAGGCACCA-3′ and antisense: 5′-CTCCTTAATGTCACGCACGATTTC-3′; DACT1: sense:5′-GCGAAGAGATGCTGGTITGT-3′ and antisense: 5′-AAATTGTGGTGTGGAGAGGG-3′. The PCR was run at 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min. Melting curve analysis was used to monitor the accumulation of the PCR products. β-actin mRNA was used as internal control to standardize the results of different mRNA expression levels. Triplicate Ct values were analyzed in Microsoft Excel using the comparative CT (▵▵CT) method as described by the manufacturer (Applied Biosystems, Foster City, CA, USA). The results were normalized against the β-actin control.
Western blot analysis
Tissue samples were homogenized in ice-cold lysis buffer (50 mmol/L Tris (pH 8.0), 100 mmol/L NaCl, 0.1% SDS, 1% NP-40, 0.5 mmol/L EDTA) containing a complete protease inhibitor cocktail (Roche, Mannheim, Germany). Proteins (20 µg) were boiled for 5 min and separated with 15% SDS-PAGE, and then transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA), blocked in 5% skim milk in phosphate buffered saline (PBS), and probed with a goat-anti-human DACT1 antibody (1:2,000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by incubation with an anti-rabbit IgG secondary antibody.
Statistical analysis
All statistical analyses were performed by SPSS version 13.0 for Windows (SPSS Inc., Chicago, IL, USA). The difference between unmethylated and hypermethylated tissues was assessed by χ2 or Fisher's exact test. A Student's t-test was used for statistical comparison in RT-PCR analysis. P < 0.05 was considered statistically significant.
RESULTS
The methylation status of the DACT1 gene
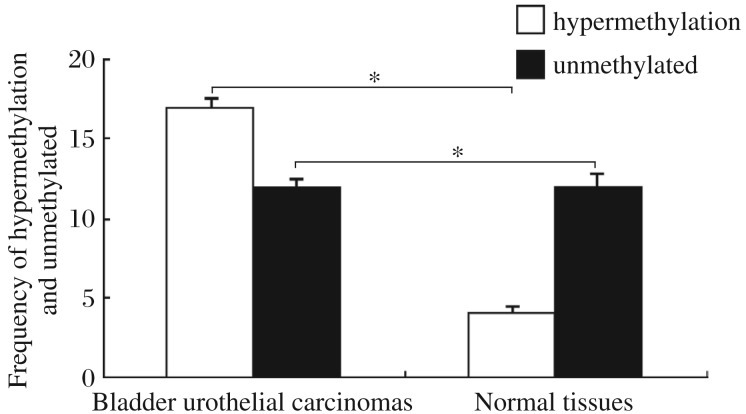
The methylation-specific PCR results showed that the DACT1 gene was mostly methylated in bladder cancer tissues (Fig. 1). The promoter of the DACT1 gene in 17 out of 29 (58.62%) cancer tissue specimens and in 7 out of 29 (25%) normal tissues was hypermethylated (P < 0.05) (Fig. 2). Thus, these data revealed that DACT1 hypermethylation may be a neoplastic feature of bladder cancers. The expression of DACT1 was low in bladder cancer tissues.
Fig. 1. The DACT1 gene is hypermethylated in bladder cancer tissues.
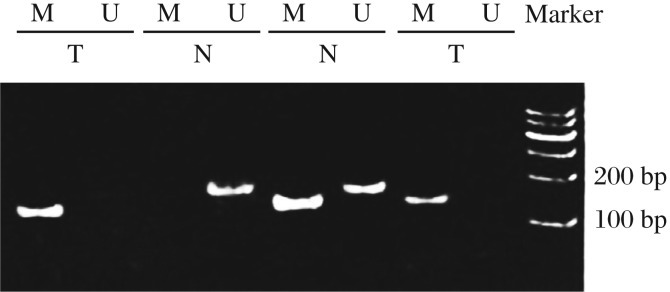
Methylation status of the promoter region CpG island of the DACT1 gene in bladder cancer and normal tissues was examined by methylation-specific polymerase (MSP) chain reaction. U: unmethylated (183 bp); M: methylated (181 bp). T:tumor tissues; N: normal tissues.
Fig. 2. Methylation-specific PCR analysis of the DACT1 promoter from normal and bladder cancer tissues.
The MSP assay showed that the frequency of hypermethylaion was 25% and 58.62% in normal tissue and bladder cancer tissues, respectively. DACT1 was hypermethylated in most bladder cancer tissues, and unmethylated in normal tissues. *P < 0.05.
The expressionof the DACT1gene
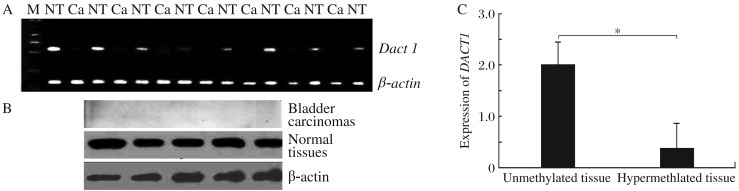
DACT1 mRNA expression was absent in most bladder carcinomas, but present in almost all normal bladder tissue (Fig. 3A). Western blot analysis confirmed that DACT1 protein was absent in bladder cancer tissues. Conversely, DACT1 protein was present in almost all normal samples (Fig. 3B). DACT1 protein was detected in some bladder cancers, but at a significantly reduced level in comparison to normal controls. The RT-PCR assay confirmed the quantitative relationship between the hypermethyaltion status and DACT1 expression (Fig. 3C): the expression of DACT1 in the hypermethylated group was lower than that in the unmethylated group (0.25±0.17 vs 0.69±0.30, P < 0.05).
Fig. 3. The expression of the DACT1.
A: RT-PCR analysis of the DACT1 expression in 29 bladder cancer samples and 29 normal bladder tissue samples. The expression of the DACT1 mRNA was low, even absent in most bladder carcinomas, but high in normal tissue. β-actin was used as an internal control. Ca: cancer tissue; NT: normal tissue. B: Western blotting analysis of DACT1 expression had a trend similar to the mRNA expression: lower expression in bladder carcinoma samples compared to normal control. DACT1 protein expression is related to hypermethylation status: lower expression is associated with hypermethylation of DACT1 gene promoter. C: RT-PCR confirms the differential expression of DACT1 mRNA between the hypermethylated and unmethylated promoter of DACT1 gene (β-actin housekeeping gene as an internal control). *P < 0.05.
Correlation between methylation status of the DACT1 gene and clinicpathological data in bladder carcinomas patients
DACT1 gene hypermethylation was closely associated with tumor size, grade and stage (P < 0.05), but not gender and age (Table 1), suggesting that the methylation rate of the DACT1 gene increased with the progression of bladder cancer.
Table 1. Correlation between methylation status of the DACT1 gene and clinicopathological characteristics in bladder urothelial carcinoma patients.
| Variables | Cases (n) | Total methylation rate (%) |
| Sex | ||
| Male | 15 | 20.6 |
| Female | 14 | 21.3 |
| P value | 0.936 | |
| Age (year) | ||
| ≤60 | 26 | 34.6 |
| > 60 | 3 | 35.3 |
| P value | 0.945 | |
| Tumor size (cm) | ||
| < 4 | 16 | 18.1 |
| ≥4 | 13 | 81.8 |
| P value | 0.016 | |
| T stage | ||
| T1/2 | 15 | 13.7 |
| T3/4 | 14 | 77.4 |
| P value | 0.021 | |
| N stage | ||
| N0 | 18 | 26.2 |
| N1 | 11 | 56.4 |
| P value | 0.029 | |
| M stage | ||
| M0 | 20 | 24.6 |
| M1 | 9 | 66.4 |
| P value | 0.032 | |
| Grade | ||
| 0 | 2 | 0 |
| I | 8 | 12.4 |
| II | 7 | 23.2 |
| III | 9 | 56.3 |
| IV | 3 | 66.6 |
| P value | 0.038 |
Data were analyzed with Fisher's exact test.
DISCUSSION
In the United States, there were approximately 68,810 newly diagnosed bladder cancer cases and 14,100 deaths were expected in 2008[16]. More than 90% of bladder carcinomas have a transitional cell origin[16]. Substantial evidence has demonstrated that many tumor suppressor genes are methylated. Shi et al.[17] found that aberrant methylation in the DBC2 promoter may be responsible for the loss of DBC2 expression in bladder cancer and this hypermethylation event could play a crucial role in the early stage of bladder tumorigenesis. Eissa et al.[18] demonstrated that methylated RARβ2 and APC are significantly higher in bladder cancer (62.8%, 59.5%) than benign tumors (16.4%, 5%), and virtually undetectable in healthy volunteers (0%) (P < 0.0001). Hypermethylation of tumor suppressor genes SFRP1 and SOCS-1 correlates with bladder carcinogenesis and development[19]. Methylation of tumor suppressor genes plays an important role in the tumorigenesis of bladder cancer. Substantial evidence has demonstrated that tumor cells display mostly hypermethylation profiles in suppressor genes, which contributes to the suppression of their expression[20].
In the present study, we first examined the hypermethylation status of DACT1 in normal and bladder cancer tissue, as well as the levels of DACT1 mRNA transcripts and protein. Hypermethylation was detected in bladder carcinomas by methylation specific PCR. The mono-alleles were detected in most bladder cancers. The DACT1 gene was hypermethylated in 58.62% (17 out of 29) urothelial carcinomas, and only 25% (7 of 29) in healthy bladder tissue (P < 0.05). These results indicate that the methylated type, which might occur either randomly or in mono-allelic manner, can be found in bladder cancer and methylation alone could repress DACT1 expression in bladder cancer. Although some cases (including normal tissues and bladder cancers) showed a hypomethylation status (methylated and unmethylated alleles), the DACT1 expression levels proved different by RT-PCR assays. Based on these findings, we hypothesize that the ratio of unmethylated DACT1 is higher, and the expression level of DACT1 may increase. In other words, the methylation status in some genes determines their expression level in tissues. Western blot analysis showed that DACT1 expression is lower in bladder cancer and the trend was the same as the hypermethylation status of the DACT1 gene. Additionally, we analyzed the correlation between methylation status of the DACT1 gene and clinicopathological features. The results suggest that increasing DACT1 methylation is correlated with tumor growth and progression. We also detected methylation in some normal bladder tissue samples, suggesting that the presence of other mechanisms, such as histone modification, may also regulate the expression of DACT1. Together with the fact that methylation of the DACT1 CpG island is common in bladder cancer, these findings indicate that DACT1 hypermethylation is related to the progression of bladder transitional cell carcinomas. Our data also suggest that other molecular mechanisms contribute to lower DACT1 expression in the tumor.
Paradoxical aberration of DNA methylation pattern is a hallmark of cancer. With a global loss of DNA methylation that coexists with regional hypermethylation of certain genes[21], it has been proposed that hypermethylation and hypomethylation in cancer are two independent processes and target different programs at different stages in tumorigenesis[22]. Hypermethylation and silencing of genes regulating proliferation are proposed to be critical for deregulating growth in early carcinogenesis, whereas hypomethylation and activation of other genes may be more important for metastasis[23]–[25]. The importance of hypermethylation and inactivation of tumor suppressor genes is well-documented in various cancers, but the role of DNA hypomethylation in advanced cancers and metastasis has been less thoroughly studied and remains hypothetical[26].
In conclusion, our study showed that hypermthylation could regulate DACT1 expression in bladder transitional cell carcinomas. From a clinical point of view, DACT1 could potentially become a target for intervention. Better understanding of the mechanism of DACT1 expression may eventually lead to novel treatment for bladder transitional cell carcinomas.
Acknowledgments
We thank Yunfei Wei, Xin Yu, and Haibing He (Department of Urology, First Affiliated Hospital of Nanjing Medical University) for critical comments and assistance in the preparation of the manuscript.
Footnotes
The authors reported no conflict of interest.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen MF, Cavenee WK. Tumor suppressors: recessive mutations that lead to cancer. Cell. 1988;53:173–4. doi: 10.1016/0092-8674(88)90376-5. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. Oncoprotein networks. Cell. 1997;88:333–46. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sincic N, Herceg Z. DNA methylation and cancer: ghosts and angels above the genes. Curr Opin Oncol. 2011;23:69–76. doi: 10.1097/CCO.0b013e3283412eb4. [DOI] [PubMed] [Google Scholar]
- 6.Arai E, Ushijima S, Fujimoto H, Hosoda F, Shibata T, Kondo T, et al. Genome-wide DNA methylation profiles in both precancerous conditions and clear cell bladder cell carcinomas are correlated with malignant potential and patient outcome. Carcinogenesis. 2009;30:214–21. doi: 10.1093/carcin/bgn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma A, Heuck CJ, Fazzari MJ, Mehta J, Singhal S, Greally JM, et al. DNA methylation alterations in multiple myeloma as a model for epigenetic changes in cancer. Wiley Interdiscip Rev Syst Biol Med. 2010;2:654–69. doi: 10.1002/wsbm.89. [DOI] [PubMed] [Google Scholar]
- 8.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol. 2010;177:21–8. doi: 10.2353/ajpath.2010.090999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murty VV, Narayan G. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:1548–9. doi: 10.1093/jnci/dji317. [DOI] [PubMed] [Google Scholar]
- 10.Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–18. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 11.Okerlund ND, Kivimae S, Tong CK, Peng IF, Ullian EM, Cheyette BN. DACT1 is a postsynaptic protein required for dendrite, spine, and excitatory synapse development in the mouse forebrain. J Neurosci. 2010;30:4362–8. doi: 10.1523/JNEUROSCI.0354-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–51. [PubMed] [Google Scholar]
- 13.Grade M, Ghadimi BM, Varma S, Simon R, Wangsa D, Barenboim-Stapleton L, et al. Aneuploidy-dependent massive deregulation of the cellular tramscriptome and apparent divergence of the Wnt/β-catenin signaling pathway in human rectal carcinomas. Cancer Res. 2006;66:267–81. doi: 10.1158/0008-5472.CAN-05-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee WC, Hough MT, Liu W, Ekiert R, Lindström NO, Hohenstein P, et al. Dact2 is expressed in the developing ureteric bud/collecting duct system of the kidney and controls morphogenetic behavior of collecting duct cells. Am J Physiol Bladder Physiol. 2010;299:740–51. doi: 10.1152/ajprenal.00148.2010. [DOI] [PubMed] [Google Scholar]
- 15.Sobin LH, Torloni H. Histologic typing of urinary bladders. In: Mostofi FK, Davis CJ, Sesterhenn IA, editors. International histological classification of tumors. 2nd edition. Geneva: World Health Organization; 1973. p. 36. [Google Scholar]
- 16.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–41. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Chen JY, Yang J, Li B, Chen ZH, Xiao CG. DBC2 gene is silenced by promoter methylation in bladder cancer. Urol Oncol. 2008;26:465–9. doi: 10.1016/j.urolonc.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Eissa S, Swellam M, El-Khouly IM, Kassim SK, Shehata H, Mansour A, et al. Aberrant methylation of RARβ2 and APC genes in voided urine as molecular markers for early detection of bilharzial and nonbilharzial bladder cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:1657–864. doi: 10.1158/1055-9965.EPI-11-0237. [DOI] [PubMed] [Google Scholar]
- 19.Chen PC, Tsai MH, Yip SK, Jou YC, Ng CF, Chen Y, et al. Distinct DNA methylation epigenotypes in bladder cancer from different Chinese sub-populations and its implication in cancer detection using voided urine. BMC Med Genomics. 2011;20:45–7. doi: 10.1186/1755-8794-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Ying J, Li J, Fan Y, Poon FF, Ng KM, et al. Aberrant promoter methylation of DLEC1, a critical 3p22 tumor suppressor for bladder cell carcinoma, is associated with more advanced tumor stage. J Urol. 2010;184:731–7. doi: 10.1016/j.juro.2010.03.108. [DOI] [PubMed] [Google Scholar]
- 21.Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci. 2010;101:36–45. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali AH, Kondo K, Namura T, Senba Y, Takizawa H, Nakagawa Y, et al. Aberrant DNA methylation of some tumor suppressor genes in lung cancers from workers with chromate exposure. Mol Carcinog. 2011;50:89–99. doi: 10.1002/mc.20697. [DOI] [PubMed] [Google Scholar]
- 23.Easwaran HP, Van Neste L, Cope L, Sen S, Mohammad HP, Pageau GJ, et al. Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus. Cancer Res. 2010;70:8015–24. doi: 10.1158/0008-5472.CAN-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding YB, Wu ZY, Wang S, Zha XM, Zheng W, Liu XA, et al. Hypermethylation of syk gene in promoter region is associated with oncogenesis, metastasis of breast carcinoma. Zhonghua Yi Xue Za Zhi (in Chinese) 2004;84:290–3. [PubMed] [Google Scholar]
- 25.Tang M, Torres-Lanzas J, Lopez-Rios F, Esteller M, Sanchez-Cespedes M. Wnt signaling promoter hypermethylation distinguishes lung primary adenocarcinomas from colorectal metastasis to the lung. Int J Cancer. 2006;119:2603–6. doi: 10.1002/ijc.22211. [DOI] [PubMed] [Google Scholar]
- 26.Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J Biol Chem. 2004;279:31735–44. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]