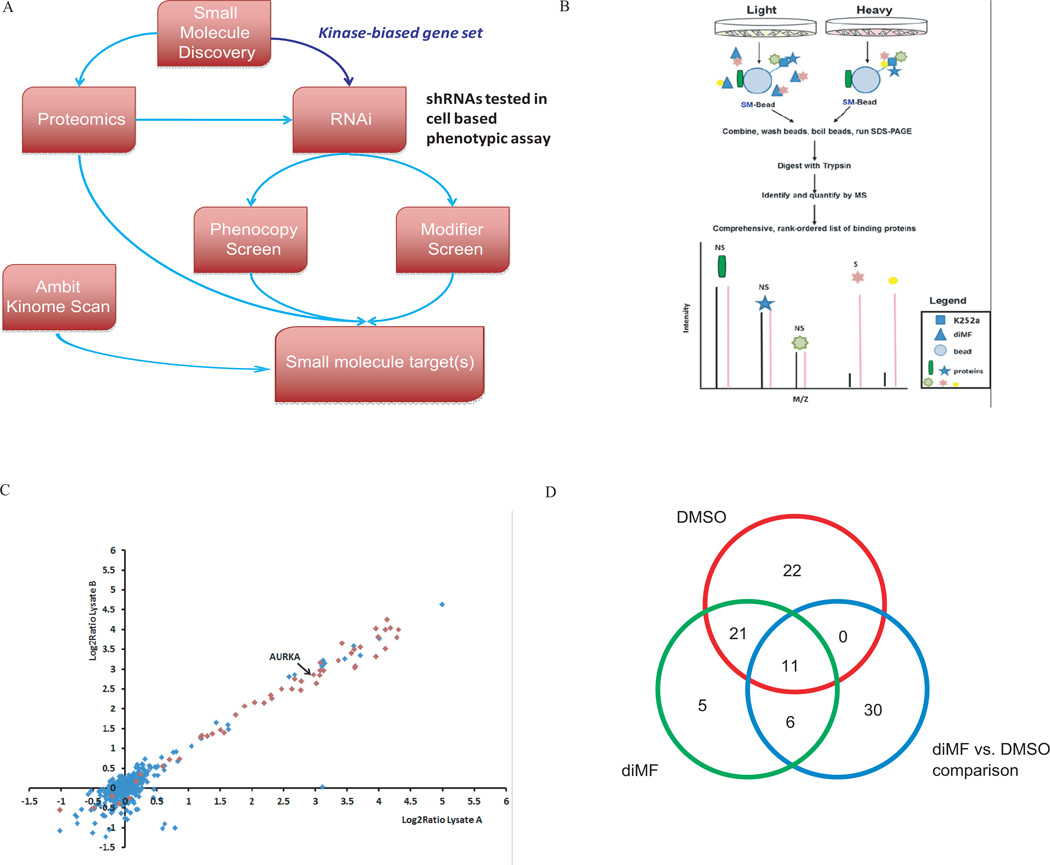
Figure 4. An integrated target ID approach identifies Aurora kinase A as a target of diMF in AMKL.
(A) Schematic representation of the integrated target identification workflow. CMK cells were transduced with shRNAs targeting the human kinome and the effects of knockdown were studied in the presence of DMSO (phenocopy screen) or a minimally effective dose of diMF at 1 µM (modifier screen). (B) Quantitative proteomic strategy for identification of specific diMF-protein interactions. Proteins in cell populations were fully metabolically labeled with light (yellow) and heavy amino acids lysine and arginine (red) using SILAC methodology. Cell lysates were incubated either with K252a-loaded beads (K252a-Beads) and excess soluble diMF competitor or K252a-Beads alone. Proteins interacting directly with diMF or via secondary and/or higher-order interactions (marked “S” for specific) were enriched in the heavy state over the light and identified with differential ratios in the mass spectrometer. Nonspecific (via binding to the bead) or K252a (NS) interactions of proteins were enriched equally in both states and have ratios close to unity. (C) Identification of significant targets of diMF using affinity proteomics with SILAC. Scatter plot of two replicate experiments of diMF at 50-fold excess over K252a on beads. Each data point is a single protein with kinases (Manning et al., 2002), represented as red diamonds and blue diamonds denoting non-kinases. Six hundred ninety-eight proteins were identified and quantified in at least three experiments, resulting in 68 proteins with a combined q-value < 0.05. (D) Venn diagram of genes scored as hits in each type of comparisons.