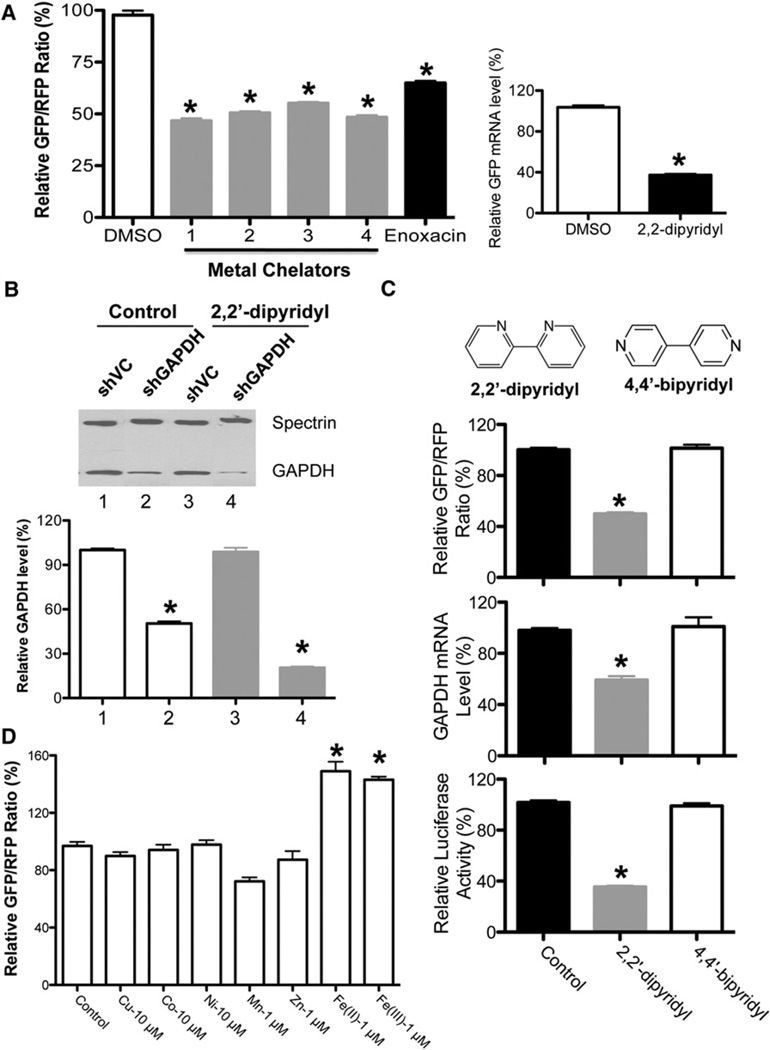
Figure 1. Identification of Iron Chelators as a Class of shRNA-Mediated RNAi Enhancer via Chemical Screen.
(A) Iron chelators enhance shRNA-EGFP-mediated gene silencing. DMSO was used as a negative control, whereas enoxacin, a previously identified RNAi enhancer, was used as a positive control. Values are mean ± SD for triplicate samples. *p < 0.001 when metal chelators were compared with control or enoxacin, and enoxacin was compared with control. The left panel shows the relative GFP/RFP ratios, while the right panel shows the GFP mRNA level determined by quantitative RT-PCR. Values are mean ± SD for triplicate samples.
(B) The metal chelator 2,2′-dipyridyl enhances shGAPDH-mediated gene silencing. The GAPDH protein levels were detected by western blots with anti-GAPDH antibody, with Spectrin used as a loading control. Metal chelator has no effect on GAPDH expression in shVC control cells. Values are mean ± SD for triplicate samples. *p < 0.001 when shGAPDH treated with 2,2′-dipyridyl was compared with shGAPDH control, and shGAPDH control was compared with shVC.
(C) The iron chelator 2,2′-dipyridyl, but not its inactive isomer 4,4′-bipyridyl, could enhance shRNA-mediated RNAi. Three reporter cell lines, shGFP, shLuciferase, and shGAPDH, were treated with 2,2′-dipyridyl and its isoform 4,4′-bipyridyl for 48 hr before analyses. Values are mean ± SD for triplicate samples. *p < 0.001 when 2,2′-dipyridyl was compared with control (DMSO) and 4,4′-bipyridyl.
(D) Iron specifically regulates the activity of the RNAi pathway. A metal substitution assay was performed with RNAi-293-EGFP/RFP reporter cells. The reporter cells were seeded in FBS-free DMEM supplemented with different metals at comparable toxicity levels for 24 hr before measuring relative fluorescence intensity. Values are mean ± SD for triplicate samples. *p < 0.001.
See also Figures S1 and S2.