Abstract
Gemin4 is a ubiquitously expressed multifunctional protein that is involved in U snRNP assembly, apoptosis, nuclear /cytoplasmic transportation, transcription, and RNAi pathways. Gemin4 is one of the core components of the Gemin-complex, which also contains survival motor neuron (SMN), the seven Gemin proteins (Gemin2–8), and Unrip. Mutations in the SMN1 gene cause the autosomal recessive disorder spinal muscular atrophy (SMA). Although the functions assigned to Gemin4 predominantly occur in the nucleus, the mechanisms that mediate the nuclear import of Gemin4 remain unclear. Here, using a novel panel of Gemin4 constructs we identify a canonical nuclear import sequence (NLS) in the N-terminus of Gemin4. The Gemin4 NLS is necessary and independently sufficient to mediate nuclear import of Gemin4. This is the first functional NLS identified within the SMN-Gemin complex.
Keywords: Gemin4, survival motor neuron, SMN, spinal muscular atrophy (SMA) nuclear localisation signal (NLS), import receptors
Introduction
Gemin4 was originally identified using a biochemical approach to identify additional members of the SMN/Gemin complex [1]. The deduced 1058 amino acid protein has a calculated molecular mass of 120kD but migrates as a 97kD protein by Western immunoblot analysis [1]. Alignment of the cDNA to the human genomic sequence indicates that Gemin4 is without introns. The international Radiation Hybrid mapping Consortium has localised Gemin4 to human Chromosome 17 and a variant of Gemin4, HCAP1, has further refined the location to 17p13.3, which is a loss of heterozygosity region in human hepatocellular carcinoma [2].
Gemin4 is part of a large protein complex, termed the Gemin complex, that also contains the spinal muscular atrophy (SMA) determining protein, survival motor neuron (SMN) [1]. SMA is the leading genetic cause of infant mortality and is characterised by loss of motor neurones in the spinal cord and severe degradation of voluntary smooth muscle, [3]. The core components of the Gemin-complex are Gemin2 (formally known as SMN-interacting protein-1, or SIP-1) [4], Gemin3 (also known as dp103/ddx20) [5], Gemin5 [6], Gemin6 [7], Gemin7 [8], Gemin8 [9] and Unrip [10]. The complex is closely associated with the splicing small nuclear ribonuceloproteins (U snRNPs) in the nucleus and the cytoplasm [11; 12; 13]. The U snRNPs are the subunits of the spliceosome and consist of a U snRNA, the Sm core proteins and several proteins unique to the various RNPs [14]. In the cytoplasm, SMN and Gemin2 perform essential roles in the assembly of the Sm core proteins on the U snRNA backbones [4; 12]. SMN and Gemin4 both interact specifically with the U snRNA and the Sm core proteins [4; 12; 15], suggesting they facilitate the assembly by forming a bridge between the two components [1; 16]. The Gemin complex is also involved in the nuclear import of the assembled U snRNPs through an interaction with the import receptor, Importin β [17]. However, no specific nuclear import sequence has been identified and confirmed in any of the complex members.
Here, using a novel panel of Gemin4 cDNA expression vectors, we have analysed the Gemin4 protein to identify cellular targeting domains. Based upon mapping experiments and in silico analysis, we have identified a nuclear localisation signal (NLS) in the N-terminus of Gemin4. We demonstrate that removal of this sequence results in the cytoplasmic accumulation of Gemin4 and that this sequence alone is independently sufficient to drive nuclear import of a heterologous protein, GFP.
Materials and Methods
cDNA Expression Constructs
Gemin4 constructs were cloned into the pEGFP (BD Bioscience), pCI-HA (Promega), and pCMV-2 (Sigma) mammalian expression vectors as described elsewhere [18], using primers listed in Table 1 and the combinations listed in Table 2. All products were amplified by PCR using commercial Gemin4 cDNA cloned into a cloning vector (RZPD; Germany) as a template. All amplified cDNA contained 5′ EcoRI and 3′ XhoI restriction sites. Cloned constructs, with their respective amino acids (aa’s) are as follows: full-length (aa 1-1058), F1 (aa 1-370), F2 (aa 300-708), F3 (aa 607-1058), F1-2 (aa 1-708), F2-3 (aa 300-1059), G4ΔN143 (aa 144-1058), G4ΔN193 (aa 194-1058), G4ΔN246 (aa 244-1058), G4ΔN293 (aa 294-1058), G4NLS (aa 194-243), G4ΔNLS (aa 1-193, 244-1058), G4Δ194-218 and G4Δ218-243.
Table 1.
Primer sequences
| Primer Name | Primer Sequence |
|---|---|
| G4 F1 | GATC GAA TTC ATG GAC CTA GGA CCC TTG AAC ATC |
| G4 F2 | GATC GAA TTC GCC AAA CTC CCC AGT GAG ACC ATT TTC |
| G4 F3 | GATC GAA TTC GAG GTA GAC CTC AGT CTG AGG ATC |
| G4 R1 | GATC CTC GAG TCA GAA GAT GCT CAT CTT CTG CAA CAG |
| G4 R2 | GATC CTC GAG TCA GAA GAT GCT CAT CTT CTG CAA CAG |
| G4 R3 | GATC CTC GAG GTT CTT CAG CAC CGT GCT CGT TTT |
| G4ΔN193 F | GATC GAA TTC GAT GAG TTC CCC CAT CCT CCA |
| G4ΔN243 F | GATC GAA TTC GCT GAC ATG CTG ACT GTG TTT |
| G4ΔN293 F | GATC GAA TTC AAG GAG GCA GAA CGG GAT GTC |
Table 2.
Primer combinations used to for each cDNA construct
| Construct | Primer Pairs |
|---|---|
| G4 FL | G4 F1 + G4 R1 |
| F1 | G4 F1 + G4 R3 |
| F2 | G4 F2 + G4 R2 |
| F3 | G4 F3 + G4 R1 |
| F1+2 | G4 F1 + G4 R2 |
| F2+3 | G4 F2 + G4 R1 |
| G4ΔN193 | G4ΔN193 F + G4 R1 |
| G4ΔN243 | G4ΔN243 + G4 R1 |
| G4ΔN293 | G4ΔN293 + G4 R1 |
Cell culture and immunohistochemistry
Human cervical carcinoma (HeLa) cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10 % (v/v) fetal calf serum (FCS) and 1 % (w/v) each of penicillin and streptomycin in 5 % (v/v) CO2 at 37 °C. Subconfluent cells were grown on coverslips and transfected with 1 μg cDNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations and as previously reported [19]. Post-transfection, cells were incubated for 4 hours in 5 % (v/v) CO2 at 37 °C, then washed and re-fed with DMEM containing 10 % FCS and incubated for a further 24 hours. Transfected cells were washed three times with 1 x phosphate-buffered saline (PBS) and then fixed with 50% acetone / 50% methanol. Immunohistochemistry staining was carried out as previously described [19; 20; 21]. Target proteins were visualised using the following antibodies: Gemin4 (C-18; Santa Cruz), Gemin2 (3-F8; Santa Cruz), Sm core protein (Y-12; Zymed), and Actin (C-2; Santa Cruz) and secondary antibodies conjugated to either FITC (green), or TRITC (red). Cell nuclei were counterstained with 4′, 6′-diamidino-2-phenylindole (DAPI).
Results and Discussion
The N-terminus of Gemin4 contains a nuclear localisation signal (NLS)
Although Gemin4 clearly localises in nuclear Cajal bodies, the mechanisms that control its nuclear import remain unclear. To determine whether Gemin4 contained a functional nuclear localisation signal (NLS), a panel of Gemin4 sub-domains was generated and transiently expressed in HeLa cells (Fig. 1). In initial experiments, full-length Gemin4 was over expressed in HeLa cells (Fig. 2). Unlike endogenous Gemin4, which is found in the cytoplasm and the nucleus, transiently expressed HA-tagged full-length Gemin4 was predominantly nuclear (Fig. 1). Similar distribution patterns were also seen with GFP and FLAG fusion tag constructs, suggesting this altered distribution is not due to the HA-tag. As all three tags are N-terminal, it is possible that the addition of any N-terminal tag alters the distribution of Gemin 4. Although HA-tagged EWS displays a similar distribution pattern when over expressed in HeLa cells, it is possible that nuclear accumulation could be a consequence of transient over expression [19]. However, although the expressed Gemin4 does not accumulate in the cytoplasm, both the FLAG and HA-tagged Gemin4 co-localised with endogenous SMN and Sm core proteins in Cajal bodies in the nucleus, demonstrating that the tags do not prevent the correct nuclear targeting of Gemin4 (data not shown).
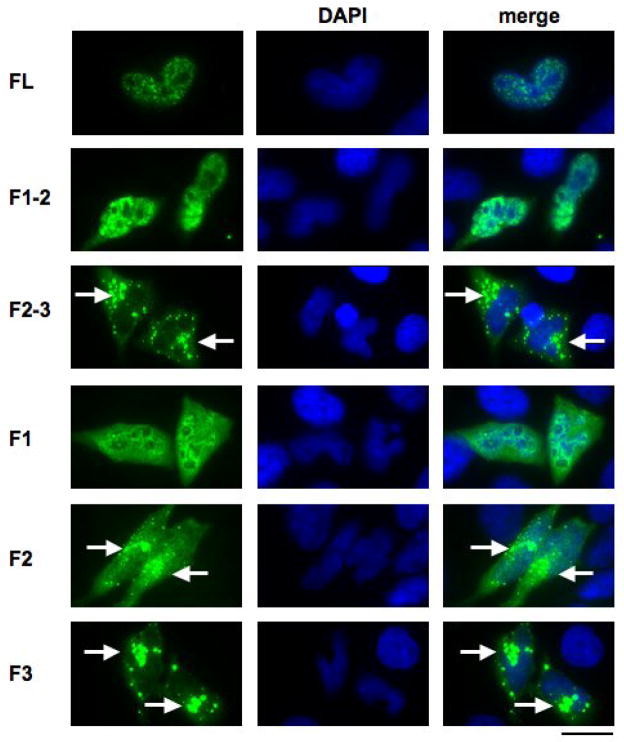
Figure 1. The N-terminal region of Gemin4 contains a nuclear localisation signal (NLS).
Sub-confluent HeLa ATCC cells (adenocarcinoma) were grown on coverslips and transiently transfected with pCI-HA containing FL-Gemin4 (aa 1-1058), F1 (aa 1-395), F2 (aa 306-708), F3 (aa 665-1058), F1-2 (aa 1-708), F2-3 (aa 306-1058). Expressed proteins, containing the HA-tag, were visualised using an anti-HA mouse monoclonal and an anti-mouse FITC-conjugated secondary antibody. All experiments were repeated with constructs cloned into the pEGFP and pFLAG-cmv vectors (data not shown). Nuclei were counterstained with DAPI (blue). Cytoplasmic aggregates formed in cells expressing cells lacking F1 (F2-3, F2 and F3) are indicated (white arrows).
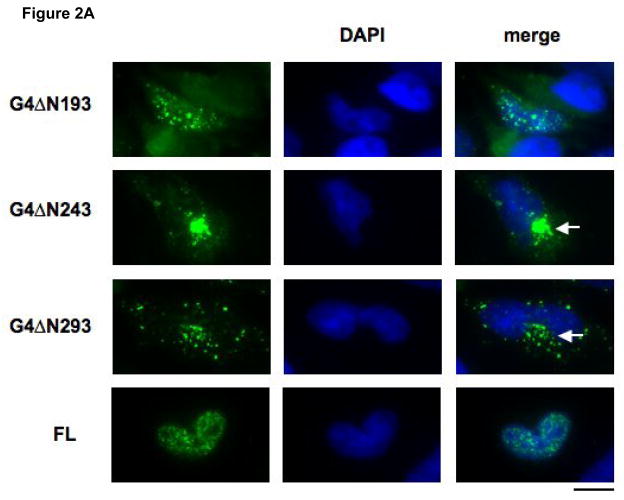
Figure 2. Identification of a canonical NLS encoded by amino acids 194-243 of Gemin4.
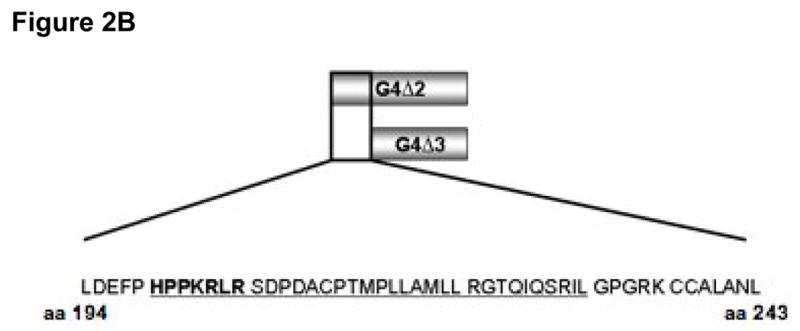
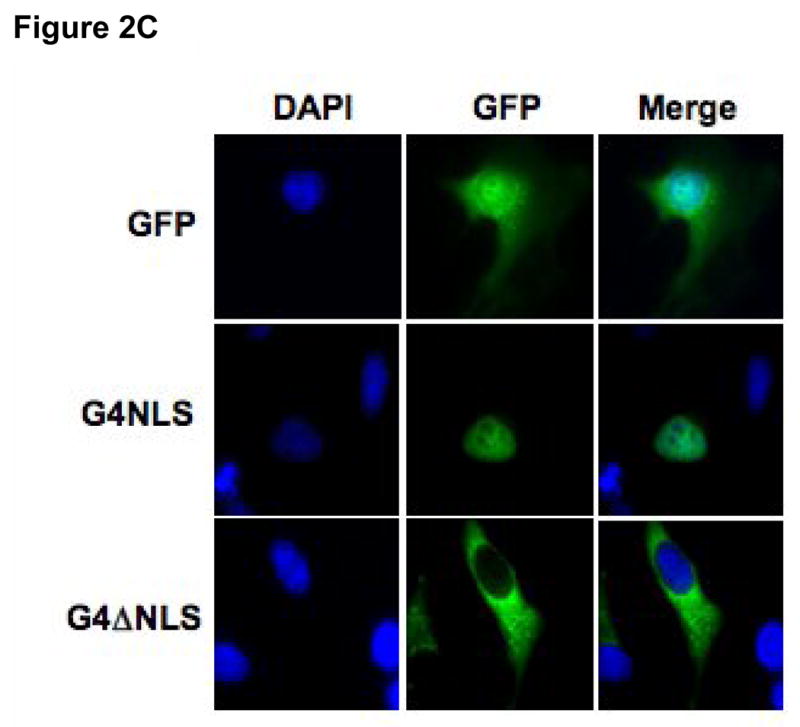
A) Sub-confluent HeLa cells were transiently transfected with 0.5 μg of a panel of truncated Gemin4 constructs: G4ΔN193, G4ΔN243, or G4ΔN293, cloned into the pCI-HA vector. Transfected cells were incubated for 24 hrs and expressed proteins were visualised using an anti-HA mouse monoclonal and an anti-mouse FITC-conjugated secondary antibody. Full-length Gemin4 (FL) is shown as a reference and is taken directly from Fig. 1. Cytoplasmic aggregates are indicated (white arrows). The bar represents ~30μm. B) A schematic of the region containing the predicted NLS. The encoded amino acids are shown, with the putative SV40-like simple (bold font) and the canonical complex (underlined) NLS’s indicated. The corresponding amino acid numbers are shown. C) Expression of aa 194-243 of Gemin4 (G4NLS) is sufficient for nuclear localisation. Removal of the predicted NLS from Gemin4 (G4ΔNLS) results in exclusive localisation of Gemin4 to the cytoplasm. G4NLS and G4ΔNLS are cloned into the pEGFP vector (BD Biosciences). An empty pEGFP vector (GFP), is shown as a reference. DAPI was used to counter-stain nuclei (blue). The bar represents~30μm.
As Gemin4 is efficiently imported into the nucleus, a panel of deletion constructs were then used to identify potential NLS’s. Constructs lacking F1 (F2, F3, and F2-3) were predominantly present within the cytoplasm with Gemin4 primarily concentrated within large cytoplasmic aggregates (Fig. 1; white arrows). The formation of these aggregates is likely a result of improper localisation of Gemin4 after the NLS has been removed. Conversely, proteins containing the amino-terminal F1 domain (F1, F1-2 and FL Gemin4; Fig. 2) localised to the nucleus (FL) or to the nucleus and the cytoplasm (F1, F1-2). Since Gemin4 sub-constructs lacking the F1 region were not detected within the nucleus, these results suggested that the F1 domain might contain a functional nuclear localisation signal (NLS) (Fig. 1). Similar distribution patterns were seen for constructs expressing N-terminal FLAG or GFP tags (data not shown).
To delineate the putative NLS encoded within the F1 domain, HA epitope tagged Gemin4 constructs with progressively shorter N-terminal domains were transiently transfected into HeLa cells: G4ΔN143 (aa 144-1058; data not shown), G4ΔN193 (aa 194-1058; Fig. 2A), G4ΔN246 (aa 244-1058; Fig. 3A), G4ΔN293 (aa 294-1058; Fig. 2A). G4ΔN143 and G4ΔN193 displayed similar distribution patterns to full-length Gemin4 (Fig. 2A). However, G4ΔN243 and G4ΔN293 localised predominantly in cytoplasmic aggregates (Fig. 2A; white arrows), suggesting that the region between G4ΔN193 and G4ΔN243 (aa 194-243) encoded the NLS required for targeting Gemin4 to the nucleus. This localisation pattern was observed with GFP-tagged and HA-tagged constructs (Fig. 2A and data not shown).
Figure 3. The SV40-like and basic regions of the conical NLS are both essential for efficient nuclear import.
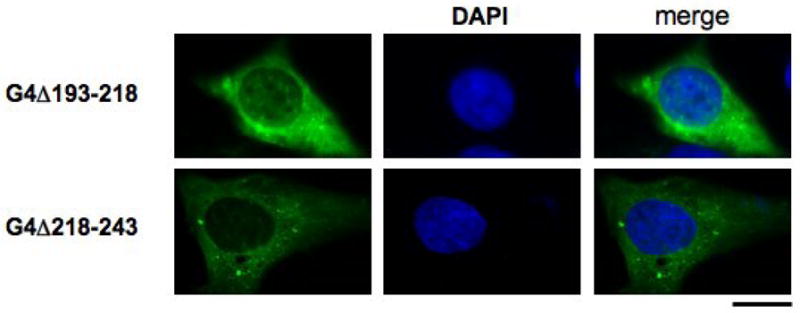
cDNA constructs lacking the SV40-like sequence (G4Δ193-218) and the basic region (G4Δ218-243) were cloned into the pEGFP vector and transiently expressed in sub-confluent HeLa cells. Nuclei were counterstained with DAPI (blue). Bars represent ~30μm.
There are three main sub-classes of NLS’s: (1) Simian Virus (SV40)-like signals, which contain a stretch of basic residues; (2) bipartite signals, which consist of two clusters of basic aminoacids separated by a spacer; and (3) Mata2-like signals, which contain a KIPIK-motif [22; 23; 24; 25; 26]. Sequence analysis of Gemin4 revealed two putative nuclear localisation signals; a strong SV40-like NLS signal (Fig. 2B; bold type) and a bipartite NLS signal consisting of the potential SV-40 signal, a 15 amino acid spacer sequence containing numerous proline residues, followed by a cluster of basic residues (Fig. 2B; underlined). Consistent with our previous studies, the region between amino acids 194-243 contained these two predicted nuclear localisation signals (Fig. 2B; bold font). To examine the requirement for the predicted Gemin4 NLS, additional Gemin4 GFP-fusion constructs were generated and transiently expressed in HeLa cells (Fig. 2C). Expression of minimal NLS sequence was independently sufficient to drive nuclear import of GFP (NLS; Fig. 2C), while its removal from Gemin4 (Gemin4ΔNLS; Fig. 2C) resulted in exclusive cytoplasmic localisation of the GFP-tagged construct. Approximately 99% of transfected cells displayed a similar distribution pattern (data not shown).
To determine whether the complete bipartite import signal was required for nuclear localisation, additional Gemin4 constructs were generated that lacked either the N-terminal (G4Δ193-218) or C-terminal (G4Δ218-243) region of the identified NLS. Both of these constructs were predominantly cytoplasmic when expressed in HeLa cells (Fig. 3). These studies suggest that both elements of the bipartite signal are required for Gemin4 nuclear localisation (Fig. 3). The N-terminal amino acids preceding the NLS did not display a localisation pattern different from GFP-alone (data not shown), confirming the specificity of the NLS sequences and further supporting the N-terminal deletion analysis that identified Gemin4 aa’s 194-243 as those responsible for nuclear localisation (Fig. 2 and 3). Therefore, the Gemin4 NLS is necessary and sufficient to target Gemin4 to the nucleus.
To identify the mechanisms that control Gemin4 import, various import receptors (snurportin-1, β1-karyopherin, β2-karyopherin, and α2-karyopherin) were immunoprecipitated using commercial antibodies to see if they associated with Gemin4. However, endogenous Gemin4 was not precipitated in a complex with any of the isolated import receptors (data not shown).
This reflects the transient nature of the nuclear import complex. In a previous report, β2-karyopherin (β2-Importin) has been identified as the import receptor that mediates the nuclear import of SMN [17]. This report did not demonstrate a complex between endogenous β2-karyopherin and SMN, and is therefore consistent with this current study. However, in the previous study an association between SMN and β2-Importin was seen using transiently over-expressed SMN with a myc tag, or in ex vivo pull down experiments using GST-tagged SMN [17].
To conclude, a nuclear localisation signal in Gemin4, encoded by amino acids 194-243, has been identified that is necessary for efficient nuclear import of Gemin4. Furthermore, this sequence is independently sufficient to mediate the nuclear import of GFP. This is the first active NLS that has been confirmed within the Gemin complex. One of the major functions performed by the SMN complex is the cytoplasmic assembly and subsequent nuclear import of the U snRNPs [1; 27]. Therefore, these studies suggest Gemin4, through the newly characterised NLS, may play a functional role in the nuclear import of the U snRNPs. Experiments are currently being performed to confirm this, and also to identify the mechanism through which the NLS mediates import.
Acknowledgments
P.J.Y. and R.M. were supported by fellowships from FightSMA and the Vandervell Foundation; D.S. was sponsored by FightSMA, A.G.T. was sponsored by IBCS, and A.M.D. is supported by a Research Grant from the Department of Veterinary Pathobiology. This work was funded by grants from FightSMA (M.A.L), and the National Institutes of Health (C.L.L, R01 NS41584; R01 HD054413).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–86. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K, Potter A, Baban D, Davies KE. Protein expression changes in spinal muscular atrophy revealed with a novel antibody array technology. Brain. 2003;126:2052–64. doi: 10.1093/brain/awg208. [DOI] [PubMed] [Google Scholar]
- 3.Bergin A, Kim G, Price DL, Sisodia SS, Lee MK, Rabin BA. Identification and characterization of a mouse homologue of the spinal muscular atrophy-determining gene, survival motor neuron. Gene. 1997;204:47–53. doi: 10.1016/s0378-1119(97)00510-6. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–21. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 5.Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–94. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem. 2002;277:5631–6. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 7.Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J Biol Chem. 2002;277:7540–5. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 8.Baccon J, Pellizzoni L, Rappsilber J, Mann M, Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J Biol Chem. 2002;277:31957–62. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- 9.Carissimi C, Saieva L, Baccon J, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J Biol Chem. 2006;281:8126–34. doi: 10.1074/jbc.M512243200. [DOI] [PubMed] [Google Scholar]
- 10.Carissimi C, Baccon J, Straccia M, Chiarella P, Maiolica A, Sawyer A, Rappsilber J, Pellizzoni L. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 2005;579:2348–54. doi: 10.1016/j.febslet.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 11.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–24. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–9. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 13.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–9. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 14.Meister G, Hannus S, Plöttner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, Fischer U. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20:2304–14. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bühler D, Raker V, Lührmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–7. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 16.Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol. 2002;22:6533–41. doi: 10.1128/MCB.22.18.6533-6541.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum Mol Genet. 2002;11:1785–95. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morse R, Shaw DJ, Todd AG, Young PJ. Targeting of SMN to Cajal bodies is mediated by self-association. Hum Mol Genet. 2007;16:2349–58. doi: 10.1093/hmg/ddm192. [DOI] [PubMed] [Google Scholar]
- 19.Young PJ, Francis JW, Lince D, Coon K, Androphy EJ, Lorson CL. The Ewing’s sarcoma protein interacts with the Tudor domain of the survival motor neuron protein. Brain Res Mol Brain Res. 2003;119:37–49. doi: 10.1016/j.molbrainres.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Young PJ, Day PM, Zhou J, Androphy EJ, Morris GE, Lorson CL. A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J Biol Chem. 2002;277:2852–9. doi: 10.1074/jbc.M108769200. [DOI] [PubMed] [Google Scholar]
- 21.Young PJ, Jensen KT, Burger LR, Pintel DJ, Lorson CL. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J Virol. 2002;76:3892–904. doi: 10.1128/JVI.76.8.3892-3904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibata S, Matsuoka Y, Yoneda Y. Nucleocytoplasmic transport of proteins and poly(A)+ RNA in reconstituted Tpr-less nuclei in living mammalian cells. Genes Cells. 2002;7:421–34. doi: 10.1046/j.1365-2443.2002.00525.x. [DOI] [PubMed] [Google Scholar]
- 23.Zakaryan RP, Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J Mol Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 24.Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–74. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honsei N, Ikuta T, Kawana K, Kaneko Y, Kawajiri K. Participation of nuclear localization signal 2 in the 3′-ETS domain of FLI1 in nuclear translocation of various chimeric EWS-FLI1 oncoproteins in Ewing tumor. Int J Oncol. 2006;29:689–93. [PubMed] [Google Scholar]
- 26.Szebeni A, Mehrotra B, Baumann A, Adam SA, Wingfield PT, Olson MO. Nucleolar protein B23 stimulates nuclear import of the HIV-1 Rev protein and NLS-conjugated albumin. Biochemistry. 1997;36:3941–9. doi: 10.1021/bi9627931. [DOI] [PubMed] [Google Scholar]
- 27.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci USA. 2005;102:17372–7. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]