SUMMARY
Tail-anchored (TA) proteins access the secretory pathway via posttranslational insertion of their C-terminal transmembrane domain into the endoplasmic reticulum (ER). Get3 is an ATPase that delivers TA proteins to the ER by interacting with the Get1-Get2 transmembrane complex but how Get3’s nucleotide cycle drives TA protein insertion remains unclear. Here, we establish that nucleotide binding to Get3 promotes Get3-TA protein complex formation by recruiting Get3 to a chaperone that hands over TA proteins to Get3. Biochemical reconstitution and mutagenesis reveal that the Get1-Get2 complex comprises the minimal TA protein insertion machinery with functionally critical cytosolic regions. By engineering a soluble heterodimer of Get1-Get2 cytosolic domains, we uncover the mechanism of TA protein release from Get3: Get2 tethers Get3-TA protein complexes into proximity with the ATPase-dependent, substrate-releasing activity of Get1. Lastly, we show that ATP enhances Get3 dissociation from the membrane, thus freeing Get1-Get2 for new rounds of substrate insertion.
INTRODUCTION
Eukaryotic cells employ a variety of sophisticated targeting pathways to efficiently and accurately deliver newly synthesized proteins to organelles (Cross et al., 2009). Membrane proteins are particularly difficult to target because their hydrophobic membrane spanning regions are prone to aggregation and must be shielded from the aqueous environment of the cytosol until they are inserted into the membrane (Wickstrom et al., 2011). The signal recognition particle (SRP) pathway solves this problem by allowing ribosomes to directly transfer membrane spanning segments into the Sec61 protein translocation channel from which they partition into the endoplasmic reticulum (ER) membrane (Rapoport, 2007).
Cotranslational targeting by the SRP pathway, however, is not an option for membrane proteins that have their ER targeting information contained in a single transmembrane domain (TMD) near the C-terminus (Kutay et al., 1995; Yabal et al., 2003). Instead, these tail-anchored (TA) proteins are posttranslationally targeted for insertion into the ER because their TMD emerges from the ribosome exit tunnel after protein synthesis is already completed. Once insertion has occurred, TA proteins are sorted to their resident organelles by vesicular transport (Kutay et al., 1995). Some prominent examples of this class of membrane proteins include most of the SNAREs, which mediate vesicular transport, a component of the Sec61 translocon, and enzymes required for ubiquitination of misfolded ER proteins. In total, there are hundreds of TA proteins in the secretory pathway of mammalian cells and they play critical roles in a variety of cell biological processes.
The GET (Guided Entry of TA proteins) pathway is the dominant cellular mechanism for handling newly synthesized TA proteins destined for the secrotory pathway. It comprises a membrane targeting and a membrane insertion stage. A recent biochemical study of the targeting stage of the GET pathway in budding yeast revealed that a TA protein chaperone called Sgt2 is the first to recognize C-terminal TMD signals with ER-targeting information (Wang et al., 2010). Sgt2 is in a complex with Get4 and Get5 (Chang et al., 2010; Ito et al., 2001; Krogan et al., 2006; Liou et al., 2007; Wang et al., 2010), which enable efficient transfer of TA proteins from Sgt2 to Get3 (Wang et al., 2010). Get3 is an ATPase chaperone that links the targeting and insertion stages of the GET pathway by delivering TA proteins for insertion into the ER membrane (see below). In mammalian cells, a complex containing the mammalian homologs of Get4 and Get5, as well Bat3, a protein that lacks an apparent yeast homolog, captures nascent TA proteins on the ribosome and delivers them to TRC40, the mammalian Get3 homolog (Mariappan et al., 2010). Collectively, these and other findings (Chang et al., 2010; Chartron et al., 2010; Leznicki et al., 2010) have revealed a conserved posttranslational targeting mechanism that enables efficient formation of Get3-TA protein complexes while staving off the potential for TA protein aggregation in the cytosol.
By comparison, the membrane insertion stage of the GET pathway is less well understood. A large-scale study of genetic interactions in yeast predicted that Get1 and Get2, two ER membrane proteins, function in the same pathway as Get3 (Auld et al., 2006; Costanzo et al., 2010; Schuldiner et al., 2005; Schuldiner et al., 2008). Further supporting this view, a large fraction of cellular Get3 is associated with the ER membrane as part of a complex with Get1 and Get2 (Jonikas et al., 2009; Vilardi et al., 2011). Therefore, Δget1/2 ER membranes are thought to be defective for TA protein insertion because they can’t recruit Get3-TA protein complexes (Schuldiner et al., 2008). It remains unknown, however, whether Get1 and Get2 comprise the minimal membrane insertion machinery of the GET pathway or whether they function to anchor Get3-TA protein complexes to the ER membrane while other membrane components carry out additional steps necessary for TA protein insertion (Borgese and Fasana, 2011).
The mechanistic study of the membrane insertion stage of most targeting pathways is complicated by the fact that insertion is coupled to extensive protein translocation across the lipid bilayer. Insertion of TA proteins, however, typically involves translocation of very short polypeptide sequences following their C-terminal TMD. Thus, the GET pathway isolates the mechanistic problem of chaperoning transmembrane domains into lipid bilayers from the complexities of protein translocation. Moreover, in contrast to cotranslational insertion by the SRP pathway, which is coupled to the elaborate ribosomal machinery, Get3 appears to be the only soluble factor required for the insertion stage of the GET pathway (Bozkurt et al., 2009; Favaloro et al., 2008; Wang et al., 2010). Like most other protein targeting systems, however, the GET pathway is also fueled by metabolic energy (Favaloro et al., 2008; Stefanovic and Hegde, 2007). Attempts to exploit the GET pathway to gain a deeper mechanistic understanding of how cells overcome the kinetic barrier to inserting transmembrane domains into lipid bilayers have been hampered by the lack of defined in vitro systems for studying the precise roles of Get1, Get2, and the nucleotide cycle of Get3.
In the present study, we identify the mechanistic basis of three nucleotide-driven steps in the GET pathway, which occur before, during, and after TA protein insertion. First, we show that nucleotide binding promotes Get3-TA protein complex formation. A key component of this mechanism is the nucleotide-dependent recruitment of Get3 to the Sgt2-Get5-Get4 complex, which delivers TA proteins to Get3. Second, we demonstrate that TA proteins insert robustly into proteoliposomes with purified Get1-Get2 transmembrane complex. Mutagenesis and binding studies identify critical functional interactions between Get3 and the cytosolic domains of Get1 and Get2. Remarkably, an engineered, soluble heterodimer of Get1 and Get2 cytosolic domains induced TA protein release from Get3 in an ATPase-dependent manner. This is achieved by Get2 recruitment of Get3-TA protein complexes into proximity with the substrate-releasing activity of Get1. Lastly, ATP binding stimulates Get3 dissociation from the membrane, thus freeing Get1-Get2 for recruitment of new substrates.
RESULTS
Nucleotide Binding to Get3 Facilitates Formation of Get3-TA Protein Complexes
The role of nucleotide in the formation of Get3-TA protein complexes remains unclear. Coexpression of Get3 and Sec22, a SNARE TA protein, in bacteria resulted in Get3-Sec22 complex formation (Yamagata et al., 2010). Under these conditions, Get3-Sec22 complex formation persisted in the presence of a nucleotide-binding mutation in Get3 (Yamagata et al., 2010). Moreover, a similar bacterial coexpression study with TRC40 (the mammalian Get3 homolog) also concluded that TRC40-TA protein complex formation is nucleotide independent (Favaloro et al., 2010). These studies, however, do not exclude a role for nucleotide under more physiological conditions in which TA proteins are delivered to Get3/TRC40 by the TMD-recognition complex (TRC) (Mariappan et al., 2010; Wang et al., 2010), which in yeast comprises Sgt2-Get5-Get4.
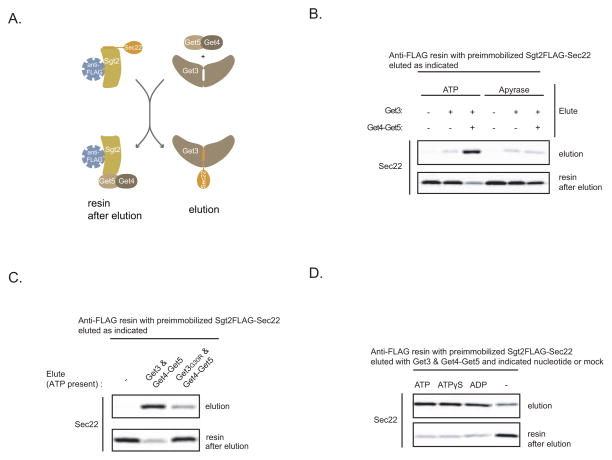
As a starting point for this study, we asked if nucleotide is required during TA protein handoff from the TRC to Get3. For this purpose we utilized our previously established in vitro system that monitors this step in the GET pathway (Figure 1A) (Wang et al., 2010). Briefly, we in vitro translate Sec22-encoding mRNA in the SGT2FLAG Δget3/5 extract and then immobilize the resulting Sgt2FLAG-Sec22 complexes on anti-FLAG resin. Addition of recombinant Get3 and Get4-Get5 (a complex formed by bacterial coexpression of Get4 and Get5) results in efficient Sec22 elution from the resin in the form Get3-Sec22 complexes that are insertion competent. Get4-Get5 doesn’t promote Sec22 elution on its own; rather it enhances the formation of Get3-Sec22 complexes by tethering Get3 to Sgt2 (Wang et al., 2010). Notably, these original experiments were carried out in the presence of an ATP regenerating system. When we used ATP instead, we observed similar results as before: Get3 alone resulted in some increase in Sec22 elution above background (Figure 1B; 2% vs. 9%) and elution was strongly stimulated upon further addition of Get4-Get5 (Figure 1B; 9% vs. 63%). On the other hand, when we excluded ATP from this assay and added Apyrase to hydrolyze any residual ATP/ADP to AMP, Sec22 elution by Get3 alone was not affected (Figure 1B: 9% with ATP vs. 10% without) but the stimulatory effect of Get4-Get5 became marginal (Figure 1B; 63% with ATP vs. 16% without). To confirm that nucleotide binding to Get3 is necessary for optimal Get3-TA protein complex formation, we used Get3 with a G30R mutation in the Walker A motif that abolishes nucleotide binding in many related ATPases (Saraste et al., 1990). Indeed, Get3G30R (Figure S1A) eluted Sec22 poorly compared to wild-type Get3 when Get4-Get5 and ATP were present (Figure 1C).
Figure 1. ATP Binding to Get3 Stimulates TA Protein Transfer from Sgt2-Get5-Get4 to Get3.
(A) Schematic of the in vitro assay for studying TA protein handoff from Sgt2 to Get3. Sec22 is a SNARE TA protein. Sgt2-Sec22 immobilized on anti-FLAG resin comes from immunoprecipitation of the SGT2FLAG Δget3/5 extract with in vitro translated Sec22-encoding mRNA. Sgt2FLAG purified in the absence of Get5 lacks Get4, (see Wang et al., 2010 for more details).
(B) In vitro translation of Sec22-encoding mRNA in the SGT2FLAG Δget3/5 extract in the presence of 35S-labeled methionine was followed by anti-FLAG immunoprecipitation (IP) and elution with the indicated Get proteins (Get3: 64 ng/μL; Get4-Get5: 96 ng/μL) or mock treatment for 20 minutes at room temperature. ATP (4 mM) or Apyrase (1 unit/μL) was also included during elution, as indicated. Following centrifugation, elutions were collected, the resin washed, and eluted again, but this time with gel loading buffer (resin after elution). The elutions were resolved by SDS-PAGE and analyzed by autoradiography.
(C) Sec22 elution from preimmobilized Sgt2-Sec22 with either Get3 or Get3G30R in combination with Get4-Get5 or mock treatment was carried out as in (B). ATP (4 mM) was included in all elutions.
(D) Sec22 elution from preimmobilized Sgt2-Sec22 with Get3 and Get4-Get5 in the presence of the indicated nucleotides (4 mM) or mock nucleotide treatment was carried out as in (B).
Next, we examined if ATP hydrolysis is required for Get3-Sec22 complexes formation under these conditions but found that it wasn’t because Sec22 elution was still efficient in the presence of ADP or ATPγS, a slowly-hydrolyzing ATP analog (Figures 1D and S1B). A recent structural study has shown that ATP and ADP both enhance Get3 binding to Get4-Get5 (Chartron et al., 2010). We have confirmed that the same is true for ATPγS (Figure S1C) and conclude that Get4-Get5 and nucleotide binding to Get3 work together to promote Get3-TA protein complex formation; in the absence of either, Get3 recruitment to Sgt2-held TA proteins is compromised.
The Get1-Get2 Transmembrane Complex is the Minimal ER Membrane Machinery for TA Protein insertions
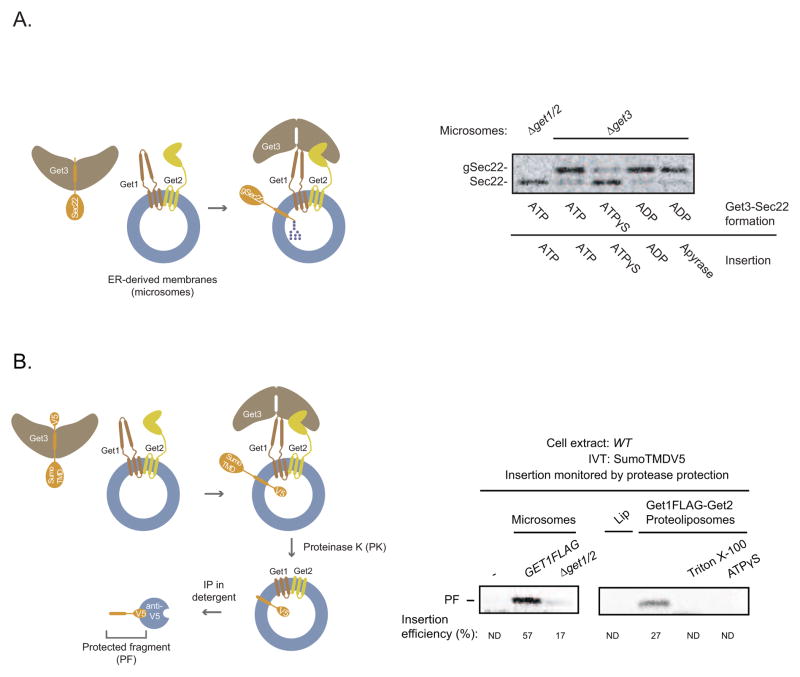
To determine if ATP hydrolysis is required during the insertion stage of the GET pathway, we first purified Get3-Sec22 complexes following Sec22 elution from Sgt2 in the presence of different nucleotides (Figure S2A). Next, we monitored Sec22 insertion into ER-derived membranes (microsomes). To facilitate detection of inserted Sec22, we used Sec22 with an N-glycan acceptor site after the TMD (Schuldiner et al., 2008). Since the glycosylation machinery of microsomes is restricted to the lumen, only inserted Sec22 will become glycosylated. As expected, we found that Get3ATP-Sec22 (superscript indicates the name of the nucleotide used during Sec22 elution from Sgt2) inserted efficiently into microsomes in a Get1/2-dependent manner (Figure 2A). By comparison, the insertion competence of Get3ATPγS -Sec22 was relatively poor (Figure 2A). Surprisingly, Get3ADP-Sec22 also inserted efficiently under these conditions and this was observed even when Apyrase was added during insertion (Figure 2A).
Figure 2. The Get1-Get2 Transmembrane Complex is the Minimal ER Membrane Machinery for TA Protein insertions.
(A) (ON THE LEFT) Schematic showing glycosylation upon microsomal insertion of Sec22, which contains a carboxyl-terminal Opsin tag with an N-glycan acceptor site. (ON THE RIGHT) Sec22 eluted with Get3His and Get4-Get5 in the presence of the indicated nucleotides (3 mM) as in Figure 1D was immunoprecipitated with anti-His resin, washed, and eluted with imidazole (Figure S2A). Elutions were incubated with Δget3 microsomes (to eliminate any endogenous Get3 that remains stably bound to Get1-Get2 during microsome preparation) or Δget1/2 microsomes for 30 minutes at room temperature in the presence of the indicated nucleotides (4 mM) or Apyrase (5 units/μL). Samples were resolved by SDS-PAGE and analyzed by autoradiography. The positions of Sec22 and glycosylated Sec22 (gSec22) are indicated.
(B) (ON THE LEFT) Schematic of protease protection assay for monitoring TA protein insertion. In vitro synthesis of SumoTMD with a C-terminal V5 epitope was followed by incubation with membranes containing Get1-Get2 and then treatment with proteinase K (PK). Following PK inactivation, digested membranes were solubilized with detergent (1% Triton) and subjected to immunoprecipitation (IP) with anti-V5 resin to detect the protected transmembrane domain fragment (PF) (see Figure S2C). (ON THE RIGHT) Wild-type (WT) extract with in vitro translated (IVT) SumoTMDV5-encoding mRNA was incubated with the indicated microsomes, liposomes (Lip), Get1FLAG-Get2 proteoliposomes, or mock incubated for 30 min at room temperature. Samples were then treated with proteinase K (PK) treatment, resolved by SDS-PAGE, and analyzed by autoradiography. Where indicated, ATPγS (3mM) and Triton X-100 (1%) were included at the time of proteoliposome or PK addition, respectively. Insertion efficiency is defined as the percentage of the protected fragment (PF) signal relative to the full-length signal prior to PK addition (not shown). Both signals were normalized for their methionine content. ND: not detected. Note that the version of SumoTMDV5 used here has a mutation in the N-glycan acceptor site to allow for direct comparison of the PF between microsomes (which would otherwise have caused the PF to become glycosylated) and proteoliposomes.
To gain a mechanistic understanding of these nucleotide effects, we developed a biochemically defined in vitro system for studying TA protein insertion. Our starting point was the observation that Get3 recruitment to the membrane is dependent on Get1 and Get2, two ER membrane proteins that form a complex with Get3 (Jonikas et al., 2009; Schuldiner et al., 2008). To determine if Get1 and Get2 comprise the minimal machinery for TA protein insertion, we first prepared microsomes from a Δget3 strain overexpressing both Get1FLAG and untagged Get2. Following detergent solubilization and anti-FLAG affinity purification, we obtained biochemical amounts of highly pure, stoichiometric Get1FLAG-Get2 complex (Figure S1A and S2B). Next, we generated proteoliposomes (Figure S2B) by removing detergent from the purified Get1FLAG-Get2 complex in the presence of synthetic phospholipids whose composition approximated that of the ER (Matsuoka and Schekman, 2000). Finally, we established a protease protection assay for monitoring TA protein insertion into proteoliposomes (Figure 2B). Here, we appended a C-terminal V5 epitope to SumoTMD, a model TA protein in which the TMD of Sec22 is fused to the C-terminus of Sumo (Wang et al., 2010). As proof of concept, we incubated microsomes with extracts containing in vitro translated SumoTMDV5-encoding mRNA and treated the samples with Proteinase K (PK). Anti-V5 immunoprecipitation revealed the presence of the desired protected fragment (PF) corresponding to the membrane inserted TMD (Figure S2C). Strikingly, we also observed the same PF when we monitored SumoTMDV5 insertion into Get1FLAG-Get2 proteoliposomes but not empty liposomes (Figure 2B). Three additional lines of evidence strongly argue that we have faithfully reconstituted the minimal insertion step of the GET pathway. First, the PF disappeared when we added detergent to the proteoliposomes during PK treatment (Figure 2B), consistent with the expectation that insertion into the proteoliposome membrane confers protease protection. Second, ATPγS inhibited insertion into proteoliposomes (Figure 2B), as it did into microsomes. Lastly, our proteoliposomes were ~25% as active as microsomes containing comparable amounts of Get1FLAG (Figure S2D; note that half of the Get1FLAG-Get2 membrane complexes are most likely reconstituted with the incorrect membrane topology). For comparison, proteoliposomes reconstituted with ER membrane components for cotranslational protein translocation were 15–20% as active as native membranes (Gorlich and Rapoport, 1993). Taken together, these data demonstrate that Get1 and Get2 are the minimal membrane machinery required for TA protein insertion into the ER membrane.
Cooperative Binding of Get3 to the Cytosolic Domains of Get1 and Get2 Enables TA Protein Insertion
Defining the mechanistic roles of Get1 and Get2 during the insertion stage of the GET pathway is complicated by the fact that the protein level of Get1 is reduced in cells lacking Get2 and vice versa (Figure S3A and Schuldiner et al., 2008). Thus, we sought to identify point mutations in Get1 and Get2 that abolish TA protein insertion without perturbing the integrity of the Get1-Get2 complex. Sequence alignment of Get1 homologs revealed that the most conserved residues reside in the predicted cytoplasmic coiled coil domain between the first two transmembrane helices (Figures 3A and S3B). Similarly, the most conserved region of Get2 is a cluster of charged residues in the predicted N-terminal cytoplasmic domain (Figures 3B and S3C). We will now present a systematic biochemical analysis, which established that TA protein insertion by Get3-TA protein complexes depends on physical interactions between Get3 and the cytoplasmic domains of Get1 and Get2.
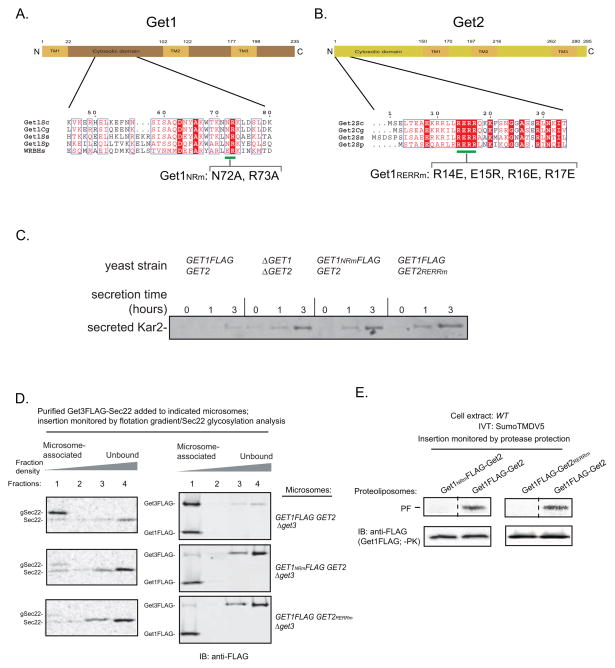
Figure 3. The Conserved Cytosolic Domains of Get1 and Get2 are Required to Recruit Get3-TA Protein Complexes for Insertion into the ER Membrane.
(A) Schematic of Get1 topology with the residues bracketing the predicted transmembrane (TM) helices and the cytosolic domain indicated. Shown below is the 45–80 amino acid region of Saccharomyces cerevisiae Get1 aligned using Clustal W2 with Get1 homologs from Candida glabrata, Scheffersomyces stipitis, Schizosaccharomyces pombe, and the Homo sapiens tryptophan basic protein (WRB). ESPript 2.0 was used to highlight identical (white in color, boxed with red), well-conserved (red in color, boxed in white) residues. The green line indicates the conserved positions that were mutated to alanines in Get1NRm.
(B) Schematic of Get2 topology with the residues bracketing the predicted transmembrane (TM) helices and the cytosolic domain indicated. Shown below is the 1–34 amino acid region of Saccharomyces cerevisiae Get2 aligned using Clustal W2 with several other Get2 fungal homologs described in (A). ESPript 2.0 was used to highlight residues as in (A). The green line indicates the conserved positions that were mutated to oppositely charged residues in Get2RERRm.
(C) The indicated strains were grown to mid-log (OD600 0.6–0.8), washed, and shifted to fresh growth media at a starting OD600 ~0.5. At the indicated times, media samples were removed, TCA precipitated, and analyzed by SDS-PAGE and immunoblotting (IB) with the anti-Kar2 antibody.
(D) In vitro translation of Sec22-encoding mRNA in the GET3FLAG extracts supplemented with additional 32 ng/μL Get3FLAG and 48 ng/μL Get4-Get5 to enhance Get3FLAG-Sec22 complex formation (data not shown). Following anti-FLAG immunoprecipitation (IP) and 3xFLAG peptide elution, elutions were split and incubated with the indicated microsomes for 30 minutes at room temperature. Samples were then overlayed with an Optiprep gradient and subjected to ultracentrifugation. Proteins were precipitated from each fraction and analyzed by autoradiography and immunoblotting (IB) with anti-FLAG. (E) Insertion of SumoTMDV5 into the indicated proteoliposomes was monitored by protease protection as in Figure 2B. Note that the proteoliposome samples were also analyzed prior to proteinase K (PK) treatment by SDS-PAGE analysis followed by immunoblotting (IB) with anti-FLAG antibody.
First, we engineered yeast strains expressing from the endogenous genomic loci either Get1 or Get2 with point mutations in their conserved cytoplasmic domains (Get1NRm: N72A, R73A; Get2RERRm: R14E, E15R, R16E, R17E). Importantly, these mutations did not change the cellular protein levels of Get1 and Get2 (Figure S3A) but resulted in cellular hypersecretion of Kar2, an in vivo hallmark of disrupted TA protein targeting by the GET pathway (Schuldiner et al., 2008), which was comparable to the phenotype of Δget1Δget2 cells (Figure 3C). Next, we prepared microsomes from these mutant strains and monitored Sec22 insertion by purified Get3-Sec22 complexes. In addition, we carried out a flotation analysis that separates microsomes from unbound Get3-Sec22 complexes. Control microsomes efficiently inserted Sec22 and recruited the majority of Get3 to the membrane (Figure 3D). In contrast, the insertion activity of Get1NRm and Get2RERRm microsomes was severely compromised consistent with their poor ability to recruit Get3 to the membrane (Figure 3D). Importantly, biochemical purification revealed that the Get1NRm-Get2 and Get1-Get2RERRm complexes are intact (Figure S1A) but defective for TA protein insertion when reconstituted into proteoliposomes (Figure 3E).
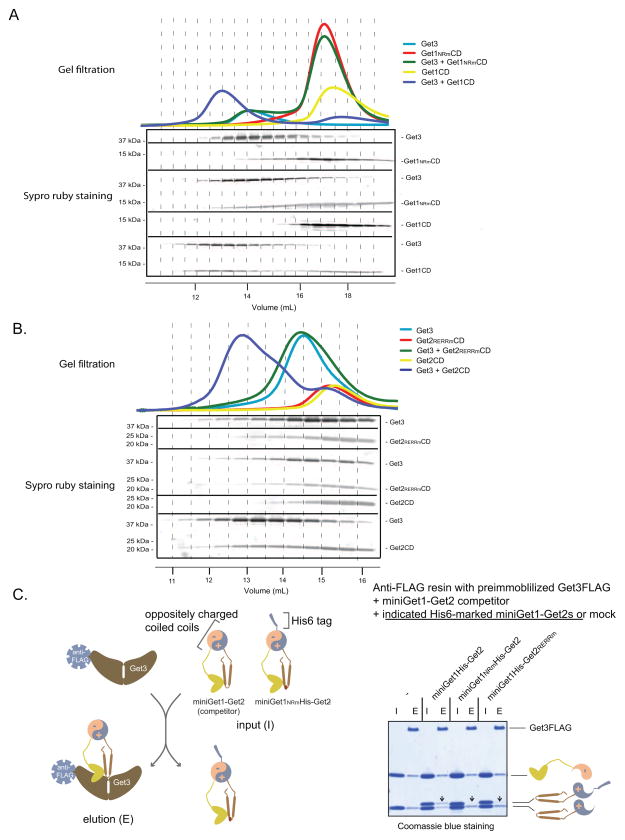
The observation that Get1NRm and Get2RERRm microsomes are unable to efficiently recruit Get3 to the membrane prompted us to test if Get3 binds to the cytoplasmic domains (CDs) of Get1 and Get2. Therefore, we expressed and purified recombinant wild-type and mutant versions of these domains (Figure S1A) and monitored Get3 binding by gel filtration analysis. We detected complex formation between Get3 and Get1CD but not Get3 and Get1NRmCD (Figure 4A). Similarly, Get3 bound to Get2CD and this interaction was disrupted by mutations in the RERR motif of Get2 (Figure 4B). Taken together, these data argue that Get3-TA protein complexes are recruited for insertion into the ER membrane by binding to the cytoplasmic domains of Get1 and Get2.
Figure 4. Cooperative Binding of Get3 to the Cytosolic Domains of Get1 and Get2.
(A) and (B) The indicated proteins were analyzed by gel-filtration chromatography (Superdex 200 10/300 GL). The relevant fractions (indicated by dash lines connecting the top A280 traces with the elution volumes on the bottom) were resolved by SDS-PAGE and visualized by Sypro ruby staining (in the middle).
(C) (ON THE LEFT) Schematic of miniGet1-Get2 binding competition assay illustrating that the miniGet1NRm-Get2 is outcompeted by the wild-type for binding to Get3 (see gel on the right). (ON THE RIGHT) Anti-FLAG resin with pre-immobilized Get3FLAG was incubated with the indicated His-marked miniGet1-Get2s and miniGet1-Get2 competitor inputs (I) (Get3FLAG:miniGet1His-Get2:miniGet1-Get2 molar ratio equals 1:2:2) in the presence of ADP (3 mM) for 20 minutes at room temperature. Following washing, the resin was eluted (E) with FLAG peptide. Samples were resolved by SDS-PAGE and visualized by Coomassie blue staining. Arrows point to the positions on the gel at which we expect to see His-tagged Get1 bands if they are able to compete for binding to Get3.
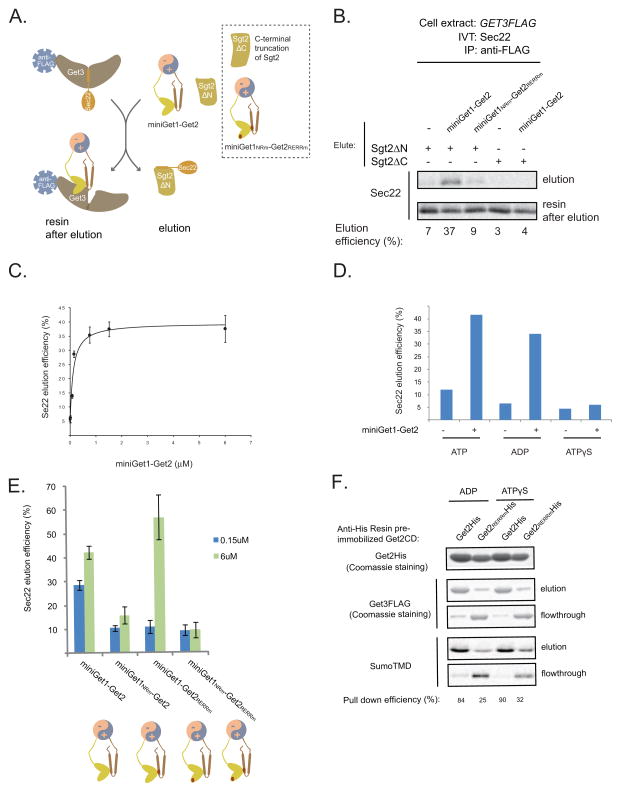
In the context of the ER membrane, the cytoplasmic domains of Get1 and Get2 are simultaneously presented to Get3-TA protein complexes. We sought to mimic this situation while still maintaining the domains in a soluble state. Since gel filtration analysis revealed no detectable binding between the Get1CD and the Get2CD (data not shown), we fused them to engineered, oppositely charged alpha helical sequences that to form a highly stable, parallel coiled coil (Moll et al., 2001). Next, we coexpressed the CD fusions and purified the resulting recombinant heterodimer, which we call miniGet1-Get2 (Figure S1A). To test if miniGet1-Get2 juxtaposes the CDs comparably to the Get1-Get2 transmembrane complex, we examined the effect of Get1NRm and Get2RERRm mutations on Get3 binding in this new soluble context. However, when we individually preimmobilized His-tagged wild-type and mutant miniGet1-Get2s on anti-His resin and monitored Get3 binding, we observed that they all robustly pulled down Get3 (Figure S4A). We suspected, however, that any reduction in Get3 binding affinity conferred by NRm and RERRm mutations in miniGet1-Get2 might be masked by the high protein concentrations used in our pull-down assay. To overcome this technical limitation, we immobilized Get3FLAG on anti-FLAG resin and monitored binding of His-tagged miniGet1-Get2s in the presence of the wild-type miniGet1-Get2 competitor from which we have removed the His tag. In this way, we could distinguish on an SDS-PAGE gel the competitor from the constructs that we were testing. As a control we established that equimolar amounts of the wild-type miniGet1His-Get2 and wild-type miniGet1-Get2 competitor bound equally well to Get3 (Figure 4C). In contrast, miniGet1NRm-Get2 and miniGet1-Get2RERRm were outcompeted for Get3 binding under the same conditions (Figure 4C). By raising the concentration of mutant miniGet1-Get2 constructs, we established that the RERRm mutations resulted in approximately a five-fold decrease in Get3 binding affinity while miniGet1NRm -Get2 was still efficiently outcompeted even at a five-fold molar excess to the wild-type competitor (Figure S4B). These data demonstrate that miniGet1-Get2 is an excellent tool for dissecting the mechanism by which the cytoplasmic domains of Get1 and Get2 work together to enable TA protein insertion.
Get1-Get2 Induces TA Protein Release from Get3 by a Dual Mechanism
We postulated that the binding of cytosolic domains of Get1-Get2 to Get3 would at least passively facilitate insertion by bringing Get3-TA protein complexes into proximity with the ER membrane. A more intriguing hypothesis is that these interactions also engender an active mechanism for releasing TA proteins from Get3. To test this, we developed an assay for monitoring Sec22 release from Get3FLAG-Sec22 immobilized on anti-FLAG resin. We reasoned that any Sec22 released in the absence of membrane would either rebind Get3 or become aggregation prone. To get around this potential problem, we included in this release assay an excess of recombinant SgtΔN, which we call TA trap (Figure 5A). We have previously shown that this deletion mutant of Sgt2 binds the TMD of Sec22 but lacks the N-terminal domain that enables it to interact with Get3 in the presence of Get4-Get5 (Wang et al., 2010). As a control, we established that very little Sec22 was eluted from immobilized Get3 in the presence of the TA trap alone (Figure 5B), arguing that TA proteins are normally tightly bound to Get3. Strikingly, addition of miniGet1-Get2 resulted in robust, dose-dependent Sec22 elution (Figures 5B and 5C). Four additional lines of evidence strongly argue that this assay monitors a key step in the TA protein insertion mechanism. First, miniGet1NRm-GetRERRm failed to release Sec22 above background (Figure 5B). Second, ATPγS efficiently blocked Sec22 release, whereas release still occurred in the presence of ADP (Figure 5D), consistent with the effects of these nucleotides on TA protein insertion. Third, released Sec22 quantitatively co-immunoprecipitated with the TA trap (Figure S5A). Fourth, replacing the TA trap with Sgt2ΔC, an Sgt2 deletion mutant that lacks the C-terminal domain, which binds TA proteins (Wang et al., 2010), resulted in no apparent Sec22 release (Figure 5B). In sum, these data strongly argue that the cytosolic domains of Get1 and Get2 enable TA protein insertion by stimulating substrate release from Get3.
Figure 5. In Vitro Reconstitution Reveals a Dual Mechanism by which Get1 and Get2 Stimulate TA Protein Release from Get3.
(A) Schematic showing release of Sec22 from immobilized Get3 by miniGet1-Get2.
(B) In vitro translation (IVT) of Sec22-encoding mRNA in the GET3FLAG extract supplemented with additional recombinant Get3FLAG (32 ng/μL) to enhance Get3FLAG-Sec22 complex formation (data not shown). After anti-FLAG immunoprecipitation (IP), the washed resin was incubated with miniGet1-Get2s (6 μM), Sgt2ΔN (0.1μg/μL), and Sgt2ΔC (0.1 μg/μL), as indicated, for 20 minutes at room temperature. Following centrifugation, elutions were collected and the resin washed and eluted again, but this time with gel loading buffer (resin after elution). The elutions were resolved by SDS-PAGE and analyzed by autoradiography. Percentage of Sec22 eluted under these conditions (elution/[elution+resin after elution]x100%) is indicated at the bottom.
(C) Dose-dependent elution of Sec22 from immobilized Get3 at different concentrations of miniGet1-Get2 (starting at 0.01 μM) in the presence of Sgt2ΔN (0.1 μg/μL). Samples were prepared and analyzed as in part (B). Average Sec22 elution efficiency and standard deviation from two independent experiments are plotted as a function of miniGet1-Get2 concentration.
(D) Sec22 elution from immobilized Get3 in the absence or presence of miniGet1-Get2 (6 μM) and Sgt2ΔN (0.1 μg/μL) with the indicated nucleotides (2.75mM) present. Samples were prepared and analyzed as in part (B).
(E) Sec22 elution from immobilized Get3FLAG-Sec22 by the indicated miniGet1-Get2s (0.15 or 6 μM) in the presence of ATP (2.75 mM). Elution was carried out in the presence of Sgt2ΔN (0.1 μg/μL) and analyzed as in Figure 5B. Average Sec22 elution efficiency and standard deviation from two independent experiments are plotted.
(F) Get3FLAG-SumoTMD was purified as described in Figure 2B and added along with additional Get3FLAG (4 μg; this reduces non-specific binding of Get3FLAG-SumoTMD to the resin, data not shown) to Ni-NTA agarose resin with preimmobilized His-tagged versions of the Get2 or Get2RERRm cytosolic domains (12 μg) for 20 min at room temperature. Following centrifugation, the flowthrough was collected and the resin was washed and then eluted with SDS gel loading buffer. Samples were resolved by SDS-PAGE and visualized by Coomassie blue staining and autoradiography.
To tease apart the individual roles of Get1 and Get2 during Sec22 release from Get3, we monitored the effects of mutations in the individual CDs on Sec22 elution by miniGet1-Get2. We found that miniGet1NRm -Get2 lost the ability to release Sec22 even at a very high concentration that is well saturating for the wild-type (Figure 5E). Further supporting this notion that the cytosolic domain of Get1 is the major source of the substrate-releasing activity of miniGet1-Get2, the Get1 cytosolic domain alone caused robust substrate release (Figure S5B). In contrast, the substrate-releasing behavior of miniGet1-Get2RERRm was concentration dependent; this mutant faired poorly at a concentration that is very near saturating for the wild-type but substrate release was rescued at a higher concentration (Figure 5E). We reasoned that higher concentrations of miniGet1-Get2RERRm are necessary to cause Sec22 release from Get3 because Get2 can no longer tether Get3-TA protein complexes into proximity with the substrate-releasing activity of Get1. Consistent with this idea, the cytosolic domain of Get2 was able to efficiently pull down Get3-TA protein complexes even in the presence of ATPγS (Figure 5F), which inhibits substrate release by miniGet1-Get2. Taken together, these data argue that Get1-Get2 uses a dual mechanism for releasing TA proteins from Get3. First, Get2 recruits Get3-TA protein complexes into proximity with Get1. Second, Get1 disrupts Get3-TA protein complexes by an ATPase-dependent mechanism.
ATP Stimulates Get3 Dissociation from the Membrane
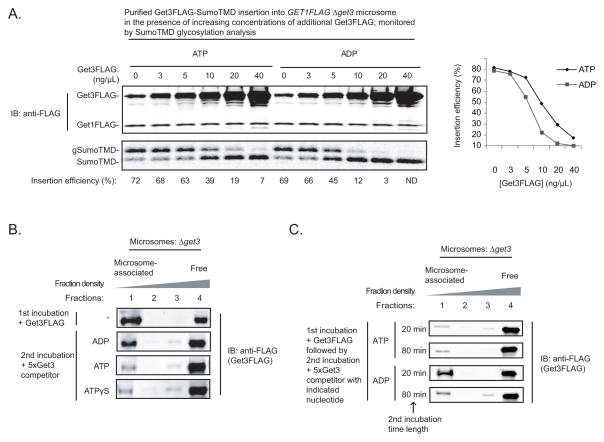
MiniGet1-Get2 caused substantial TA protein release from Get3 even in the presence of ADP, which is aligned with our earlier observation that Get3ADP-TA protein complexes insert efficiently into microsomes (Figure 2A). Why then is TA protein insertion dependent on an ATPase? Notably, the microsomes we used for our insertion experiments are prepared from a Δget3 strain. In contrast, wild-type microsomes contain abundant amounts of Get3, which copurifies as part of the Get1-Get2-Get3 complex (Jonikas et al., 2009). Thus, we reasoned that ATP would confer a more significant advantage on TA protein insertion under conditions in which Get1-Get2 accessibility is limiting. To test this, we monitored insertion of purified Get3FLAG-SumoTMD into GET1FLAG Δget3 microsomes in the presence of increasing concentrations of free Get3FLAG. As expected, in the absence of any additional Get3FLAG, SumoTMD was inserted efficiently in the presence of either ATP or ADP (Figure 6A; 72% vs. 69%). In contrast, at higher concentrations of Get3FLAG, similar to the one we have measured in cells relative to Get1FLAG (Figure S6A), SumoTMD insertion became significantly more efficient with ATP compared to ADP (Figure 6A: 39% vs. 12%). At even higher concentrations of free Get3FLAG, we detected significant insertion only in the presence of ATP (Figure 6A).
Figure 6. ATP Stimulates TA Protein Insertion when Get3 Dissociation From the Membrane is Rate-Limiting.
(A) Get3FLAG-SumoTMD was purified as in Figure 3D and incubated with the GET1FLAG Δget3 microsomes in the presence of either ADP or ATP and additional Get3FLAG, as indicated, for 30 minutes at room temperature. Samples were resolved by SDS-PAGE and analyzed by autoradiography and immunoblotting (IB) with anti-FLAG. Insertion efficiency is defined as the percentage of SumoTMD that is glycosylated (gSumoTMD/[gSumoTMD+SumoTMD]x100%). The insertion efficiencies in the presence of ADP and ATP at different concentrations of added Get3FLAG is plotted on the right. ND: not detected.
(B) Δget3 microsomes (0.8 A280 units) were incubated with Get3FLAG (0.1 μg) in the absence of nucleotide for 20 minutes at room temperature (1st incubation), and then split and incubated with the indicated nucleotides (4 mM) and Get3 competitor (0.5 μg; lacking the FLAG tag) for another 20 minutes at room temperature (2nd incubation). Samples were analyzed by flotation analysis and immunoblotting (IB) with anti-FLAG as described in Figure 3D.
(C) Δget3 microsomes (0.8 A280 units) were incubated with Get3FLAG (0.1 μg) in the absence of nucleotide for 20 minutes at room temperature (1st incubation), and then split and incubated with the indicated nucleotides (4 mM) and Get3 competitor (0.5 μg; lacking the FLAG tag) for 20 or 80 minutes at room temperature (2nd incubation). Samples were analyzed by flotation analysis and immunoblotting (IB) with anti-FLAG as described in Figure 3D.
One possible explanation for how ATP stimulates TA protein insertion under these multiple turnover conditions is that it increases the rate of Get3 dissociation from the membrane. To test this, we prebound Get3FLAG to Δget3 microsomes and then added an excess of untagged Get3 competitor, which prevents Get3FLAG rebinding to the membrane (Figure S6B), in the presence of different nucleotides. Membrane flotation analysis revealed that in the absence of nucleotide or in the presence of ADP, Get3FLAG remained tightly bound to the membrane (Figures 6B, 6C, and S6C). In contrast, when either ATP or ATPγS were present, Get3FLAG rapidly dissociated from the membrane (Figures 6B and 6C). These data argue that ATP binding promotes Get3 dissociation from the membrane following TA protein insertion, thus freeing Get1-Get2 for a new round of substrate recruitment.
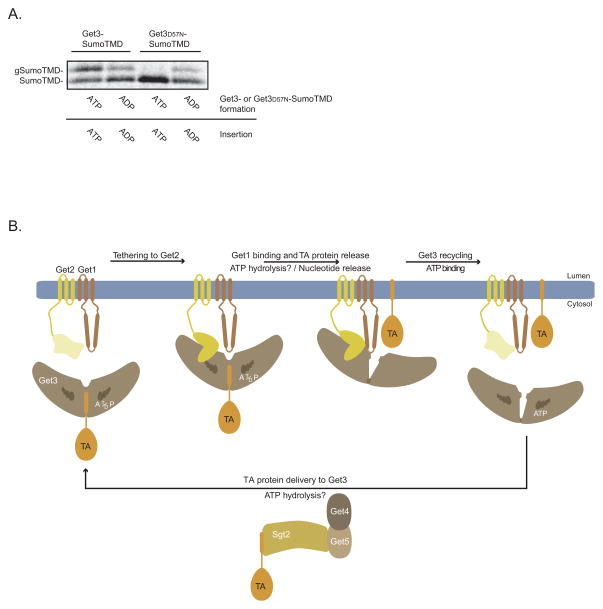
Returning to our original observation that Get3 in the ATPγS state is non-permissive for TA protein insertion while Get3 in the ADP state can only sustain a single round of the GET pathway (Figure 2A). In an effort to synthesize these phenomena, we reasoned that ATP hydrolysis by Get3 enables repeated transitions between these two states as part of the normal cellular mechanism. To test this, we used Get3 with a D57N mutation that is predicted to abolish ATPase activity by disrupting coordination of a nucleophilic water molecule. Indeed, we observed no substrate release or insertion by Get3D57NATP-SumoTMD when ATP was present (Figure 7A and S7A), while ADP actually rescued both defects (Figures 7A and S7A). Taken together, these data strongly argue that Get3’s ATPase cycle drives a molecular switch, which coordinates the targeting and insertion stages of the GET pathway.
Figure 7. The ATPase Activity of Get3 Switches the GET Pathway from the Targeting to the Insertion Stage.
A) Get3FLAG-SumoTMD and Get3D57NFLAG-SumoTMD complexes were purified from wild-type yeast cell extract as in Figure 3D with the following modification. Complexes prepared in the presence of ADP were first passed through G-50 spin columns after in vitro translation to remove nucleotides before adding ADP and recombinant proteins as in Figure 3D. Get3FLAG-SumoTMD and Get3FLAG D57N-SumoTMD were incubated with the GET1FLAG Δget3 microsomes in the presence of either ADP or ATP, as indicated, for 30 minutes at room temperature. Samples were resolved by SDS-PAGE and analyzed by autoradiography as in Figure 6A.
B) Schematic showing the role of nucleotide and GET pathway components during TA protein targeting and insertion into the ER membrane. See Discussion for more details.
DISCUSSION
Our data support the following working mechanistic model for the GET pathway (Figure 7B). ATP binding to Get3 enables efficient TA protein transfer from the Sgt2-Get5-Get4 to Get3. The resulting Get3-TA protein complexes are then recruited to the membrane by the cytoplasmic domain of Get2. This positions Get3 into proximity with the Get1 cytoplasmic domain that disrupts TA protein binding to Get3 by an ATPase-dependent mechanism. TA protein release from Get3 enables insertion into the lipid bilayer. Lastly, ATP binding to empty Get3 enhances Get3 dissociation from the Get1-Get2 complex and ushers another round of TA protein targeting.
Structural studies of Get3 (Simpson et al., 2010) provide additional support for this model. They have shown that Get3 is a flexible homodimer with a zinc finger hinge. In the nucleotide-free state, Get3 is in an open conformation that becomes closed to varying degrees upon nucleotide binding. In the most tightly closed conformation, Get3 assembles a large, composite groove with a hydrophobic character that is essential for TA protein binding. We have shown that nucleotides and Get4-Get5 work together to enhance TA protein transfer from Sgt2-Get5-Get4 to Get3. From a structural perspective, this is most likely achieved by the stabilization of a closed Get3 conformation, which increases the receptiveness of Get3 for the hydrophobic transmembrane domains of TA proteins.
Our observation that Get2 mediates tethering of Get3-TA protein complexes is consistent with a recent structural study of a Get2 N-terminal region alone and in complex with Get3 (Stefer et al., 2011). Specifically, the Get2 N-terminus is unstructured in solution but assumes alpha helical secondary structures, including a helix that spans the critical RERR motif, when it binds to a site on each Get3 subunit that is distal to the dimer interface. This coupled binding-folding phenomenon is also characteristic of the chloroplast SRP targeting system (Falk et al., 2010), suggesting unstructured domains might be a general strategy used by membrane insertion machines to efficiently explore perimembrane space for potential substrates.
Protein targeting factors have to tightly bind their newly synthesized hydrophobic substrates and shield them from aggregation in the cytosol. During the insertion stage of targeting pathways, however, targeting factors have to efficiently let go of their substrates. Our understanding of how most insertion machineries induce their targeting factors to let go of their substrates is still rudimentary. In the present study, we resolve this key conceptual problem as it pertains to the GET pathway. We were aided in this effort by the modular nature of the substrate-releasing activity of the Get1-Get2 transmembrane complex, which is contained in the cytoplasmic domain of Get1. A recent structural study of Get3 in complex with the cytosolic domain (CD) of Get1 provides a potential explanation for Get1’s substrate-releasing activity (Stefer et al., 2011). It revealed that Get1 CD is a coiled coil that sterically disrupts inter-subunit Get3 interactions necessary to form a closed Get3 dimer. In other words, Get1 CD might disrupt substrate binding by forcing a transition from a closed, high substrate affinity to an open, low substrate affinity conformation of Get3. Interestingly, Get1 CD can also form an interaction with Get3 in the closed state (Stefer et al., 2011). Establishing the precise step at which Get3 hydrolyzes ATP to enable Get1-mediated substrate release is an important future goal. Advances in loading biochemical amounts of pure, recombinant TA proteins (Leznicki et al., 2010) onto Get3 and monitoring ATP hydrolysis during the targeting and insertion steps in the GET pathway should help resolve this issue.
Lastly, how are TA proteins inserted into the lipid bilayer following their release from Get3? This could occur by an unassisted mechanism in which the released hydrophobic anchors interact with the cytosolic leaflet of the ER membrane and then insert themselves without any assistance from Get1/2/3 (Borgese and Fasana, 2011). A more complex alternative is that TA proteins released from Get3 engage an insertase mechanism that guides them into the lipid bilayer (Rabu et al., 2009; Stefanovic and Hegde, 2007). In fact, this possibility is suggested by our preliminary observation that liposomes with tethered miniGet1-Get2 on their surface are competent for TA protein release but not membrane insertion. More concretely, our work restricts the number of moving parts on a possible insertase mechanism to just three components. Furthermore, it provides a conceptual framework and the tools necessary to look for insertase-defective mutant Get1/2/3 machinery that fails to insert TA proteins into the ER membrane after they are released from Get3.
EXPERIMENTAL PROCEDURES
Plasmid and S. cerevisiae Strain Construction
Plasmid and yeast strain construction are described in the Supplemental Experimental Procedures. Strain genotypes are listed in Table S1 therein.
Native FLAG Immunoprecipitation (IP) and Elution with Get Proteins or 3xFLAG Peptide Following In Vitro Translation
See Wang et al, 2010 for a detailed description of the TA protein hand-off assay used in Figure 1. Get3FLAG-TA protein complexes were purified by native FLAG IP followed by 3xFLAG peptide elution as described previously (Wang et al., 2010).
Get1FLAG-Get2 Purification and Proteoliposome Reconstitution
Large-scale anti-FLAG affinity purification of the Get1FLAG-Get2 complex from digitonin-solubilized microsomes was carried out as described previously (Denic and Weissman, 2007). Proteoliposomes were prepared by removal of detergent from purified Get1FLAG-Get2 with SM2 Biobeads (Bio-Rad) in the presence of synthetic phospholipids as described previously (Denic and Weissman, 2007).
Microsome Flotation Analysis
Microsome flotation in an Optiprep gradient was carried out as described previously (Schuldiner et al., 2008), with minor modifications as detailed in the Supplemental Experimental Procedures.
Gel Filtration Complex Analysis
Proteins used in the experiments shown in Figures 4A and 4B were incubated at 9 μM (with the exception of Get1CD; 6 μM) each in a total volume of 200 μL SEC buffer (50 mM HEPES-NaOH pH 6.8, 150 mM NaCl, 2% glycerol, and 2mM β-mercaptoethanol) at 4° C for 30 minutes with agitation. Following centrifugation for 10 minutes at 21,000 rcf at 4° C, samples were injected onto a GE Superdex 200 10/300 GL size exclusion column.
Sec22 Release From Anti-FLAG Immobilized Get3FLAG-Sec22
2.5 μg of anti-FLAG M2 antibody (Sigma) was preimmobilized onto 0.5 mg of Protein G Dynabeads (Invitrogen) according to the manufacturer’s instructions. Each 15 μl of in vitro translations (with 32 ng/μL Get3FLAG, 48 ng/μL Get4-Get5 added during translation to enhance Get3FLAG-Sec22 complex formation) were incubated with 0.5 mg Protein G Dynabeads with preimmobilized anti-FLAG and incubated for 30 minutes at 4 °C with agitation. Following washing with ice-cold IP buffer (20 mM HEPES-KOH pH 7.4, 2 mM Mg(OAc)2, 100 mM KOAc, 2 mM DTT, 14% glycerol), 0.25 mg resin was eluted in a 36 μl final volume with recombinant proteins in IP buffer as described in the figure legends. Resin remaining after elution was washed and eluted with SDS-PAGE sample buffer as described previously for the analogous TA protein hand-off assay (Wang et al., 2010).
Supplementary Material
Table 1.
S. cerevisiae Strains Used in this Study
| Genetic Background | Deletion(s) | Epitope Tag | Strain Number |
|---|---|---|---|
| BY4741 | (none) | Get3FLAG::kan | VDY10 |
| Get1FLAG::kan | VDY9 | ||
|
Δget1::kan Δget2::nat |
(none) | VDY35 | |
| Δget3::kan | (none) | VDY36 | |
|
Δget3::ura Δget5::his |
Sgt2FLAG::kan | VDY47 | |
| Δget3::his | Get1FLAG::kan | VDY82 | |
| Get1NRmFLAG::kan | VDY84 | ||
| Get1FLAG::kan Get2RERRm | VDY85 | ||
| nat::pTDH3 Get1NRmFLAG::kan ura::pTDH3 Get2 | VDY168 | ||
| nat::pTDH3 Get1FLAG::kan ura::pTDH3 Get2RERRm | VDY173 | ||
| Obtained from a cross between BY4741 and BY4742 | Δget3::kan | nat::pTDH3 Get1FLAG::kan nat::pTDH3 Get2 | VDY343 |
Highlights.
Nucleotide binding to the Get3 ATPase promotes Get3-TA protein complex formation.
The Get1-Get2 transmembrane complex inserts TA proteins delivered to the ER by Get3.
Get2 tethers Get3-TA protein complexes for ATPase-dependent disruption by Get1.
ATP binding stimulates Get3 recycling from the membrane after TA protein insertion.
Acknowledgments
We thank V. Dotsch and I. Sinning for sharing unpublished Get1/2/3 structural data; Z. Newby for his preliminary work on Get3 binding to the cytosolic domains of Get1 and Get2; Gary Mak for generating and purifying miniGet1-Get2 constructs; E. O’Shea for allowing us the extensive use of her equipment; A. Murray, D. Kahne, S. Stefer, N. Bradshaw, and E. Feinberg for useful discussions; B. Stern, E. O’Shea, E. Feinberg, J. Weissman, and members of the Denic lab for critical reading of the manuscript; B. Toyama for generous help with graphics; V.D. wishes to thank W. Lim who impressed upon him as a student the power of studying molecular biology by engineering protein-protein interactions; Harvard University supported this work.
References
- Auld K, Hitchcock A, Doherty H, Frietze S, Huang L, Silver P. The conserved ATPase Get3/Arr4 modulates the activity of membrane associated proteins in Saccharomyces cerevisiae. Genetics. 2006:215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta. 2011;1808:937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, Wild K, Bange G, Favaloro V, Rippe K, Hurt E, Dobberstein B, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci U S A. 2009:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Chuang Y, Ho Y, Cheng M, Sun Y, Hsiao C, Wang C. Crystal structure of Get4/Get5 complex and its interactions with Sgt2, Get3 and Ydj1. J Biol Chem. 2010 doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Suloway CJ, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci U S A. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear E, Sevier C, Ding H, Koh J, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross B, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. NATURE REVIEWS MOLECULAR CELL BIOLOGY. 2009:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Denic V, Weissman J. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell. 2007;130:663–677. doi: 10.1016/j.cell.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem. 2010;285:5954–5962. doi: 10.1074/jbc.M109.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro V, Vilardi F, Schlecht R, Mayer MP, Dobberstein B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J Cell Sci. 2010;123:1522–1530. doi: 10.1242/jcs.055970. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M, Collins S, Denic V, Oh E, Quan E, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman J, et al. Comprehensive Characterization of Genes Required for Protein Folding in the Endoplasmic Reticulum. Science. 2009:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnerthilger G, Hartmann E, Wiedenmann B, Rapoport TA. TRANSPORT ROUTE FOR SYNAPTOBREVIN VIA A NOVEL PATHWAY OF INSERTION INTO THE ENDOPLASMIC-RETICULUM MEMBRANE. EMBO JOURNAL. 1995:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou S, Cheng M, Wang C. SGT2 and MVY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae. Cell Stress Chaperones. 2007:59–70. doi: 10.1379/CSC-220R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Schekman R. The use of liposomes to study COPII- and COPI-coated vesicle formation and membrane protein sorting. Methods. 2000;20:417–428. doi: 10.1006/meth.2000.0955. [DOI] [PubMed] [Google Scholar]
- Moll JR, Ruvinov SB, Pastan I, Vinson C. Designed heterodimerizing leucine zippers with a ranger of pIs and stabilities up to 10(−15) M. Protein Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: the beginning for the end? J Cell Sci. 2009:3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins S, Thompson N, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt J, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt H, Schwappach B, Weissman J. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PJ, Schwappach B, Dohlman HG, Isaacson RL. Structures of Get3, Get4, and Get5 provide new models for TA membrane protein targeting. Structure. 2010;18:897–902. doi: 10.1016/j.str.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Hegde R. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Lohr F, Bernhard F, et al. Structural Basis for Tail-Anchored Membrane Protein Biogenesis by the Get3-Receptor Complex. Science. 2011 doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrom D, Wagner S, Baars L, Ytterberg AJ, Klepsch M, van Wijk KJ, Luirink J, de Gier JW. Consequences of depletion of the signal recognition particle in Escherichia coli. J Biol Chem. 2011;286:4598–4609. doi: 10.1074/jbc.M109.081935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabal M, Brambillasca S, Soffientini P, Pedrazzini E, Borgese N, Makarow M. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J Biol Chem. 2003:3489–3496. doi: 10.1074/jbc.M210253200. [DOI] [PubMed] [Google Scholar]
- Yamagata A, Mimura H, Sato Y, Yamashita M, Yoshikawa A, Fukai S. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.