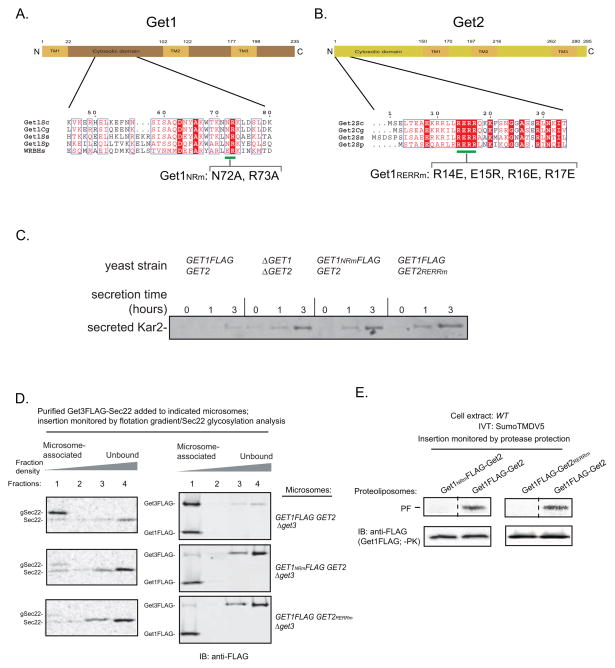
Figure 3. The Conserved Cytosolic Domains of Get1 and Get2 are Required to Recruit Get3-TA Protein Complexes for Insertion into the ER Membrane.
(A) Schematic of Get1 topology with the residues bracketing the predicted transmembrane (TM) helices and the cytosolic domain indicated. Shown below is the 45–80 amino acid region of Saccharomyces cerevisiae Get1 aligned using Clustal W2 with Get1 homologs from Candida glabrata, Scheffersomyces stipitis, Schizosaccharomyces pombe, and the Homo sapiens tryptophan basic protein (WRB). ESPript 2.0 was used to highlight identical (white in color, boxed with red), well-conserved (red in color, boxed in white) residues. The green line indicates the conserved positions that were mutated to alanines in Get1NRm.
(B) Schematic of Get2 topology with the residues bracketing the predicted transmembrane (TM) helices and the cytosolic domain indicated. Shown below is the 1–34 amino acid region of Saccharomyces cerevisiae Get2 aligned using Clustal W2 with several other Get2 fungal homologs described in (A). ESPript 2.0 was used to highlight residues as in (A). The green line indicates the conserved positions that were mutated to oppositely charged residues in Get2RERRm.
(C) The indicated strains were grown to mid-log (OD600 0.6–0.8), washed, and shifted to fresh growth media at a starting OD600 ~0.5. At the indicated times, media samples were removed, TCA precipitated, and analyzed by SDS-PAGE and immunoblotting (IB) with the anti-Kar2 antibody.
(D) In vitro translation of Sec22-encoding mRNA in the GET3FLAG extracts supplemented with additional 32 ng/μL Get3FLAG and 48 ng/μL Get4-Get5 to enhance Get3FLAG-Sec22 complex formation (data not shown). Following anti-FLAG immunoprecipitation (IP) and 3xFLAG peptide elution, elutions were split and incubated with the indicated microsomes for 30 minutes at room temperature. Samples were then overlayed with an Optiprep gradient and subjected to ultracentrifugation. Proteins were precipitated from each fraction and analyzed by autoradiography and immunoblotting (IB) with anti-FLAG. (E) Insertion of SumoTMDV5 into the indicated proteoliposomes was monitored by protease protection as in Figure 2B. Note that the proteoliposome samples were also analyzed prior to proteinase K (PK) treatment by SDS-PAGE analysis followed by immunoblotting (IB) with anti-FLAG antibody.