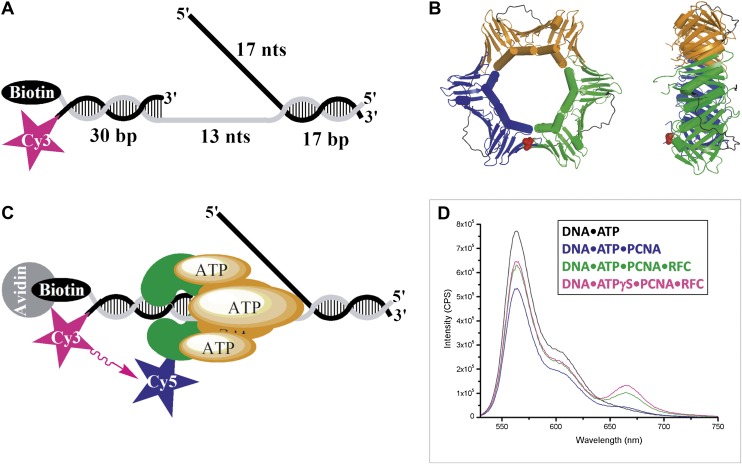
Figure 1. RFC-mediated loading of PCNA onto DNA.
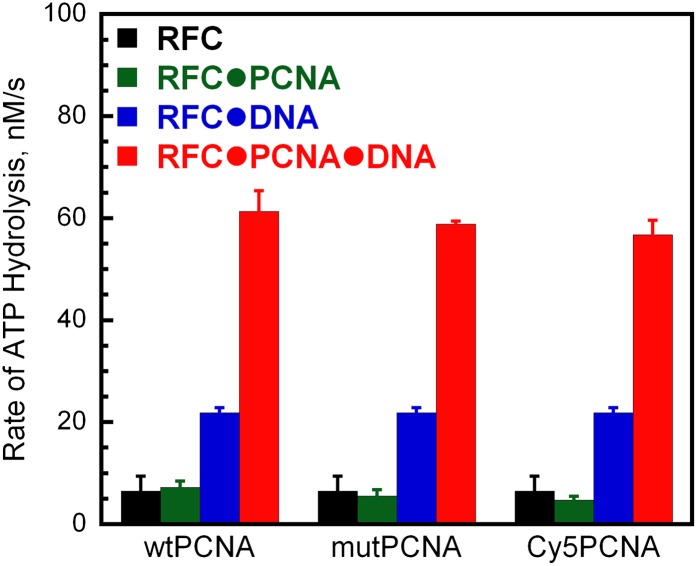
(A) Schematic representation of the Cy3-P/T DNA substrate used in this study. The primer had a Cy3 dye at the 5′ end and biotin was attached to the 3′ end of the template. Sequences for the primer, template, and flap constructs of all substrates used in this study are shown in Table 1. The recombinant human proteins used in this study are shown in Figure 1—figure supplement 1 .(B) Model of human PCNA generated using Pymol from PDB code 1AXC (Gulbis et al., 1996). PCNA subunits are shown in ribbon form in green, orange, and blue. The asparagine 107 residue, shown in red for one PCNA monomer in space-filling form, was mutated to cysteine for dye labeling. On average, each PCNA trimer has at least one labeled monomer. The mutations nor the labeling of PCNA had any effect on its ability to interact of RFC (Figure 1—figure supplement 2). Frontal and side views are shown. (C) Schematic representation of RFC-catalyzed loading of PCNA onto DNA. The N107C residue of PCNA is located on the face opposite that which interacts with RFC and faces the Cy3 FRET donor on the P/T DNA. (D) Fluorescence emission spectra obtained by exciting the Cy3-P/T DNA with a 514-nm light source. Cy5-PCNA can be excited through FRET from Cy3 only when the two dyes are in close proximity (<∼10 nm). Cy5 fluorescence intensity peaks at 665 nm.