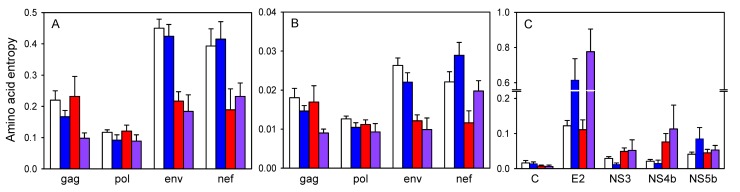
Figure 1. Association between amino acid variability and T-cell epitopes in subtype B HIV-1 (A, B) and HCV 1a (C).
Mean ± SEM entropy (H) is shown for sites not mapping to any T-cell epitopes (white) and for those mapping to TH epitopes (blue), CTL epitopes (red), or both (purple). In (A) and (C) amino acid entropy was quantified at the host population level (100 sequences from different patients), whereas in (B) it was quantified at the intrapatient level (average from 100 patients containing ≥10 sequences each). For HIV, only Gag, Pol, Env, and Nef are shown because they contain the vast majority of T-cell epitopes. No significant differences in variability associated with T-cell epitopes were found in other genes. Regions with overlapping reading frames were excluded from the analysis. For HCV, only genes with at least five sites in each category were plotted. Notice that the y-axis is broken to accommodate the extremely variable epitopes in E2.