Abstract
Unprotected amide protons can undergo fast hydrogen exchange (HX) with protons from the solvent. Generally, NMR experiments using the out-and-back coherence transfer with amide proton detection are affected by fast HX and result in reduced signal intensity. When one of these experiments, 1H-15N HSQC, is used to measure the 15N transverse relaxation rate (R2), the measured R2 rate is convoluted with the HX rate (kHX) and has higher apparent R2 values. Since the 15N R2 measurement is important for analyzing protein backbone dynamics, the HX effect on the R2 measurement is investigated and described here by multi-exponential signal decay. We demonstrate these effects by performing 15N R2CPMG experiments on α-synuclein, an intrinsically disordered protein (IDP), in which the amide protons are exposed to solvent. We show that the HX effect on R2CPMG can be extracted by the derived equation. In conclusion, the HX effect may be pulse sequence specific and results from various sources including the J coupling evolution, the change of steady state water proton magnetization, and the D2O content in the sample. To avoid the HX effect on the analysis of relaxation data of unprotected amides, it is suggested that NMR experimental conditions insensitive to the HX should be considered or that intrinsic R2CPMG values be obtained by methods described herein.
Keywords: Hydrogen exchange, intrinsically disordered protein, 15N R2 relaxation
Introduction
15N backbone transverse relaxation experiments (R2) using the CPMG (Carr–Purcell–Meiboom–Gill) pulse train (15N R2CPMG) have been used to investigate dynamics of backbone amides in globular proteins (Farrow et al. 1994; Kay et al. 1989; Mandel et al. 1995) and have also been applied to study NMR dynamics of intrinsically disordered proteins (IDPs) (Buevich and Baum 1999; Buevich et al. 2001; Eliezer 2009; Mittag and Forman-Kay 2007; Wright and Dyson 2009). 15N R2CPMG rates have been important indicators for protein backbone dynamics and have provided information about the structural mobility on the ps to ns timescale and about conformational exchange processes on the μs to ms timescale (Mittermaier and Kay 2009; Palmer III 2004; Wang and Palmer 2003). When backbone amide protons in proteins are not protected from the solvent water, those protons undergo fast hydrogen exchange (HX). Although HX rates of amide protons and R2CPMG rates of amide nitrogens are measurements of different events in protein dynamics, fast HX may affect the measurement of 15N R2CPMG rates. Since 15N R2CPMG rates are measured indirectly with 1H-15N HSQC using the out-and-back coherence transfer with a prolonged relaxation time in the pulse sequence, measured rates can be modulated due to the signal intensity of the HSQC vulnerable to fast HX. Some NMR experiments have been designed to measure fast HX rates indirectly during the 15N time evolution or relaxation. In the R1zz relaxation experiment of two spin order (NzHz) (Skrynnikov and Ernst 1999), the HX effect destroys the NzHz spin operator and this results in the combined HX and R1zz relaxation rate. In the 15N R2 relaxation experiment of the HCN-type coherence transfer (Kateb et al. 2007), the fast HX effect that dephases the 15N spin operator is correlated with the 1JNH coupling evolution. The presence or absence of 1H decoupling during the relaxation time results in a significant difference in the HX effect on 15N R2 relaxation rates. In the 15NH/D-SOLEXSY experiment using the (HACACO)NH pulse sequence, H/D exchange rates were measured during the 15N t1 evolution in H2O/D2O (50%/50%) (Chevelkov et al. 2010). Based on these experiments, it is expected that the 15N R2CPMG relaxation rate measured by the HSQC-type NMR experiments may be modulated by fast HX although the mechanism of the HX effect is different from previously reported mechanisms and includes the spin state of 15N, the J-coupling evolution, or the solvent environment.
Here we show that R2CPMG rates are modulated by fast HX rates in α-synuclein (αSyn), a protein associated with Parkinson’s disease (Chiti and Dobson 2006; Goedert 2001). If 15N R2CPMG rates are modulated by both conformational exchange and HX, it will be complicated to interpret the relaxation data without knowing the magnitude of the HX effect on the 15N R2CPMG rates. In particular, when residues with large conformational exchange are located at the solvent accessible surface areas, 15N R2CPMG rates may be a combination of both conformational exchange and fast HX rates (Bertini et al. 2002; Tugarinov and Kay 2003). αSyn is an intrinsically disordered protein (IDP) (Eliezer et al. 2001; Uversky et al. 2000; Wu et al. 2008) and forms structurally disordered ensembles in solution which possess enhanced HX rates at neutral pH and high temperature (Croke et al. 2008). We demonstrate the relationship between fast HX and the 15N R2CPMG rates at different pH states and show that the 15N relaxation rates that are affected by fast HX can be corrected quantitatively. The methods discussed here may be applied to other IDPs or partially folded proteins that are studied at neutral pH to distinguish the inherent R2 values from those associated with fast hydrogen exchange.
Materials and methods
Preparation of αSyn protein
The expression and purification of 15N-labeled αSyn was done by previously described protocols (Wu et al. 2009). Lyophilized αSyn was dissolved in phosphate buffer at pH 7.4 and 6.2 (10 mM phosphate, 137 mM NaCl, 2 mM KCl). Prior to applying NMR experiments, samples were filtered by a 100 kDa filter (Millipore) to remove invisible small aggregates. Final sample concentrations were adjusted to 250 μM with 10% (v/v) D2O for all NMR experiments.
NMR data acquisition
The 15N R2CPMG experiment was performed using a modified pulse sequence of gNhsqc from the Varian BioPack library; the main modification is an optional condition for the proton steady state (SS) pulse in the beginning of the pulse sequence (Fig. 2). The NMR data were collected twice at a proton frequency of 800 MHz and 15 °C without and with the SS pulse. Relaxation time delays for R2CPMG experiments were 0.01, 0.03, 0.05, 0.07, 0.09, 0.13, 0.17, 0.21, and 0.25 (s) at pH 7.4 and pH 6.2. The CPMG inter-pulse delay (2τ) was 1.25 ms and the recycle delay (d1) for each scan was 2 seconds. Spectral complex points were 1024 and 256 in the t2 and t1 dimensions, respectively. Relaxation data were acquired by interleaving relaxation time delays and t1 increments. Data were processed and analyzed by NMRPipe (Delaglio et al. 1995). The relaxation data were fit by two different methods to measure the apparent R2CPMG values; (1) single exponential fit as described in other previous studies (Bertoncini et al. 2007; Bussell and Eliezer 2001) and (2) multi-exponential fit introduced in this paper (discussed in results) to remove the HX effect.
Fig. 2.
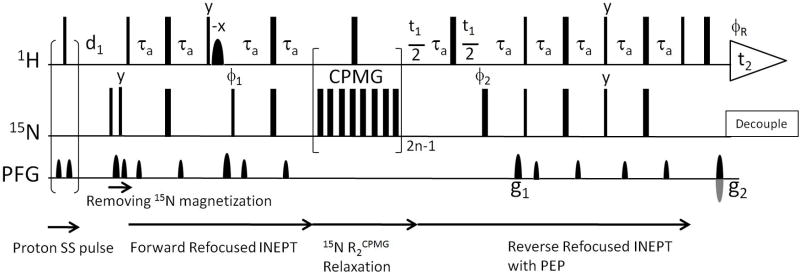
The 15N R2CPMG pulse sequence. The gNhsqc (with T2 = ‘y’) from the Varian BioPack library is outlined. 90° and 180° hard pulses are represented by narrow and wide rectangles. Pulse phases are written on the pulses but if not mentioned, phases are on the x-axis. The sinc shaped water flip-back pulse is used during the forward refocused INEPT to minimize the water saturation by bringing the water magnetization to the z-axis. Delay τa is 0.25/1JNH. Phase cycles are φ1 = x, −x, φ2 = x, x, y, y, −x, −x, −y, −y, and φR = x, −x, −x, x. To test the HX effect, an optional proton steady state (SS) pulse is introduced in the beginning of the pulse sequence. The implemented SS pulse is gradient – RF pulse – gradient where the RF pulse is a proton 90° hard pulse and the gradient is the strength of 10 gauss/cm and the duration of 0.5. 2n-1 ( = 1, 3, 5, etc) cycles of 15N CPMG pulse trains are applied for the R2CPMG measurement where the single cycle takes the 10 ms relaxation delay and a 180° proton pulse. The inter-pulse delay between 180° 15N pulses in CPMG is 1.25 ms. The gNhsqc adapts the sensitivity enhanced HSQC with PEP (Preservation and Equivalent Pathways) and the gradient coherence selection (by g1 and g2). Other experimental details are available from Varian BioPack.
To measure hydrogen exchange rates (kHX), CLEANEX (Hwang et al. 1998) experiments including a fast 1H-15N HSQC detection scheme were performed by using the pulse sequence of gCLNfhsqc from the Varian BioPack library. The following mixing times were used; 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07 (s) at pH 7.4 and 0.01, 0.02, 0.04, 0.06, 0.08, 0.1, 0.12 (s) at pH 6.2. The reference fast 1H-15N HSQC spectra were acquired to serve as V0 in the recommended fitting equation (Hwang et al. 1998). The recycle delay for each scan was 2 seconds and spectral complex points were 1024 and 256 in the t2 and t1 dimensions, respectively. Hydrogen exchange data were acquired by interleaving mixing times and t1 increments. Data were processed and analyzed by NMRPipe (Delaglio et al. 1995). The intensity build-ups by mixing times were fitted to obtain two unknown kHX and R1A, app (apparent R1 of amide protons) with a fixed R1B, app (apparent R1 of water protons, 0.6 Hz).
Results and Discussion
The 15N R2CPMG relaxation data of αSyn acquired at pH 7.4 and pH 6.2 (Fig. 1) were acquired by using the pulse sequence (Fig. 2) without the proton steady state pulse and the data were analyzed with single exponential fits as described in other previous studies (Bertoncini et al. 2007; Bussell and Eliezer 2001). The resulting 15N R2CPMG values showed that values at pH 7.4 were elevated relative to those at pH 6.2 and also showed some sequence-specific differences; specifically the N-terminal residues have slightly larger values overall than the C-terminal residues (Fig. 1). Individual R2CPMG values showed non-uniform deviations along residues and averages of reported 15N R2CPMG values were 4.21 ± 0.55 Hz at pH 7.4 and 3.54 ± 0.46 Hz at pH 6.2. The discrepancy in 15N R2CPMG values between two pH’s may arise from changes in dynamics of αSyn or by changes in conformational exchange or folding states of the protein.
Fig. 1.
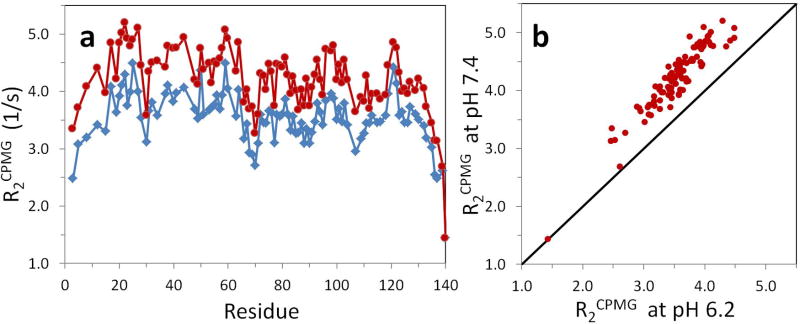
(a) Apparent R2CPMG values of αSyn at pH 7.4 (red circles) and pH 6.2 (blue rhombi) at 15°C. R2CPMG relaxation data acquired without the proton SS pulse (Fig. 2) were fit with a single exponential decay function. There are large and systematic differences between them in addition to small random deviations. (b) Correlation between R2CPMG values at pH 7.4 and pH 6.2.
However, pH-dependent water hydrogen exchange (HX) rates of amide protons may be a primary cause for large differences in 15N R2CPMG values since αSyn is known as an IDP (Eliezer et al. 2001; Uversky et al. 2000; Weinreb et al. 1996; Wu et al. 2008) and measurement of HX rates show that the protein undergoes very fast HX on the order of 5 to 20 Hz (Croke et al. 2008) and that its amide protons are not protected from H/D hydrogen exchange. In Supporting Information, HSQC spectra of αSyn acquired at various experimental conditions were similar to each other except for the residue H50 due to the pH titration of the imidazole. This supports that the intrinsic properties (including R2CPMG values) of αSyn are similar to each other at pH7.4 and pH 6.2. In this paper, we show that the pH-dependent differences in 15N R2CPMG can be derived from the pH-dependent hydrogen exchange (HX) rates. Improvement of the correlation between 15N R2CPMG values between pH 7.4 and pH 6.2 is demonstrated and 15N R2CPMG values without HX effect are obtained.
To describe the HX effect on the backbone 15N R2CPMG NMR experiment, the conventional H/D exchange (exchange between amide hydrogen and solvent deuterium) is considered as one of HX mechanisms because of the mixed solvent environment of 90% H2O and 10% D2O. For H/D exchange, it is assumed temporarily that the protein 15N spin is 100% locked in the Nx spin state during the 15N relaxation measurement period (t) and H/H exchange (exchange between amide hydrogen and solvent hydrogen) has no HX effect. The pulse sequence of the 15N R2CPMG NMR experiment using refocused HSQC is outlined in Fig. 2. The out-and-back nature of HSQC requires the NH chemical integrity, which selects the protein-NH by forward refocused INEPT and detects the same protein-NH by reverse refocused INEPT. By H/D exchange, the in-phase Nx spin state (HENx) after forward refocused INEPT can lose the attached H population (HE) and the deuterium populated DENx spin state is generated. Then, the DENx state fails to produce the NMR signal through reverse refocused INEPT.
In H/D exchange, the loss of proton occupancy on the amide is directly proportional to the D2O content in the solvent. When H2O/D2O fractions in the mixed solvent are Q/(1-Q), populations of NH/ND are Q/(1-Q) in equilibrium. However, the initial NH population selected by the forward refocused INEPT is repopulated into NH and ND by multiplying Q and (1-Q) if HX is completed during the relaxation period. Since the reverse refocused INEPT selects only the NH population, the HSQC signal is subject to signal loss by a maximum of (1-Q). By introducing the HX rate constant (kHX), the NMR signal intensity decay (I) can be a multi-exponential in equation 1 in combination with the 15N R2CPMG during the CPMG relaxation time (t).
| (1) |
Equation 1 is valid when the pseudo-first order kinetics can be applied for HX at the slow exchange regime (kHX < R2CPMG). At prolonged relaxation times under kHX > R2CPMG, HX reaches equilibrium and the backward exchange (from ND to NH) cannot be ignored in the time scale of the R2CPMG relaxation. This requires the Bloch-McConnell equation for chemical exchange (Hansen and Led 2003; McConnell 1958) which includes two different R2CPMG rates (R2NH for the protonated nitrogen and R2ND for the deuterated nitrogen) instead of a single R2CPMG in equation 1. At the extreme fast exchange (kHX ≫ R2NH), the R2CPMG (from equation 1) appears as a population-weighted average of R2NH and R2ND and the relative error of R2CPMG (from equation 1) to R2NH (from Bloch-McConnell equation) is a maximum of 5.5 % in our calculation (see Supporting Information). Especially under our experimental conditions (kHX / R2CPMG ≤ about 10 and Q = 0.9), the relative error of R2CPMG is estimated to be less than 4.5 % (see Supporting Information). Since it is difficult to obtain R2NH and R2ND separately from a single data set of a multi-exponential signal decay, equation 1 can be used as a good approximation to interpret the HX effect resulting in the large difference of R2CPMG values as shown in Figure 1.
In the case of H/H exchange happening during the 15N relaxation time period (t), the HENx state needs to be elaborated by using single element spin operators; HENx = (0.5Hα + 0.5Hβ)Nx to understand the role of the exchanged protons. By H/H exchange, an exchanging proton spin replaces Hα and Hβ with c1Hα + c2Hβ where c1 and c2 are probability coefficients from the water proton and their ensemble averages are 0.5 each. As shown in equation 2, coefficients (c1 and c2) of the exchanged H are not correlated with coefficients of 0.5 but the ensemble average generates the original HENx spin state back. If the water proton magnetization has c1 = 0.5 + δ and c2 = 0.5 − δ where δ indicates a negligible population difference, the exchanged HzNx term does not contribute to the signal through the reverse refocused INEPT. Thus the result of H/H exchange during the 15N R2CPMG relaxation has no effect on the Nx spin operator.
| (2) |
The H/H exchange needs an additional consideration which is not necessary for H/D exchange since protons are the directly observed nucleus and variable pulses and durations are applied. In the pulse sequence (Fig. 2), 180° pulses are incremented by 2n-1 with the increase of the 15N R2CPMG relaxation delay (t). Even though the experiment starts with many dummy scans to reach a steady state, we need to be careful about the relationship between the maintenance of the steady state and the increase in the R2CPMG relaxation delay. The different number of proton pulses and delays with an increase in the 15N R2CPMG relaxation delay may perturb the steady state of the proton magnetization, particularly the bulk water proton magnetization. The pulse sequence (Fig. 2) is designed to minimize the saturation of the water proton magnetization but additional pulses and delays may result in increased saturation of the water proton magnetization. With the increment in the number of 180° proton pulses, the water proton magnetization may be partially and progressively saturated. This causes a different amount of HX between amide and water protons during the fixed recycle delay (d1) when d1 is shorter than at least 5 times the water T1 relaxation time which is a typical experimental condition. It means that the steady state of the water proton magnetization can be altered depending on the 15N R2CPMG relaxation delay and it modulates the steady state of the amide proton magnetization through HX.
It will be difficult to quantify how much water proton magnetization is affected by proton pulses and delays during the 15N R2CPMG relaxation delay due to radiation damping but this effect can be removed by applying a steady state (SS) pulse (or a purge pulse) right after the acquisition time (t2) and before the recycle delay (d1). The proton SS pulse destroys the effect of the water flip-back pulse designed for minimizing water saturation (Fig. 2) and randomizes (saturates and dephases) the water proton magnetization right after acquisition. Although the SS pulse attenuates the 1H-15N HSQC signal intensity through HX when the amide hydrogen is under fast HX, it will ensure that the same amount of H/H exchange happens during the d1 delay regardless of the change of pulses and delays in the pulse sequence. For this reason, 15N R2CPMG experiments were run with and without the proton SS pulse to test HX effect. The implemented SS pulse in the 15N R2CPMG experiment is gradient - 1H 90° pulse - gradient in Fig. 2 but the other type of SS pulses such as the x-axis trim pulse followed by the y-axis trim pulse will be effective, as well.
When the in-phase Nx spin state is not locked completely by the CPMG pulses during the relaxation period, a minor population of anti-phase 15N magnetization is present by the 1JNH coupling evolution. The HX effect on the anti-phase 15N magnetization (2NyHz) will be different from the HX effect on the in-phase magnetization Nx. The H/H exchange effect on 2NyHz (equation 3) is considered in a similar way to the case of the H/H exchange of the in-phase Nx spin state (equation 2). The resulting coefficients of (c1 – c1) and (c2 – c2) by HX will become zero by ensemble averaging.
| (3) |
Since in-phase and anti-phase spin states undergo different HX effect, in-phase / anti-phase populations in the 15N R2CPMG NMR experiment need to be known and have been formulated in equation 4 through the time-averaged R2 of the exchanging in-phase and anti-phase magnetizations via J-coupling
| (4) |
where R2in and R2anti are transverse relaxation rates of in-phase and anti-phase spin states, J is the 1JNH coupling constant, and is the CPMG refocusing delay (Griesinger and Ernst 1988; Palmer III et al. 1992). Coefficients of R2in and R2anti are effective fractions of in-phase and anti-phase states; fin = 0.5{1+sinc(2πJτ)} and fanti = 0.5{1-sinc(2πJτ)}. The fraction of the anti-phase state (fanti) is estimated as 0.011 with τ = 0.625ms and 1JNH = 93Hz under the experimental conditions. Since the HX effect on 15N relaxation is included only in the anti-phase spin state, the apparent R2 (R2app) is approximated by R2CPMG + fantikHX which includes the HX effect along with equation 4. By combining this and equation 1, the resulting multi-exponential function for HX effect is in equation 5.
| (5) |
Thus the total HX effect in the 15N R2CPMG NMR experiment using refocused HSQC results from the combined effects of H/D exchange, H/H exchange, and in-phase/anti-phase J-coupling exchange. A relevant kinetic model is presented in scheme 1 and the derivation of equation 5 is in Supporting Information. In scheme 1, the total kHX was divided into QkHX and (1-Q)kHX where Q is the preserved hydrogen fraction through H/H exchange. Therefore, QkHX is the H/H exchange rate and (1-Q)kHX is the H/D exchange rate in the mixed solvent of H2O and D2O. The multi-exponential fit of the R2CPMG relaxation data with five parameters (I0, Q, fanti, kHX, and R2) shown in equation 5 is not plausible but can be reduced to a fit with two unknown parameters (I0 and R2) and with three known Q, fanti, and kHX parameters. Since Q (= 0.9) and fanti (= 0.011) are estimated from the H2O content and the anti-phase fraction under the controlled experimental condition, kHX rates need to be measured separately.
Scheme 1.
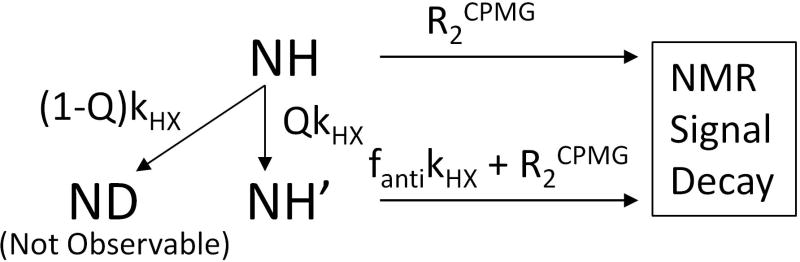
Hydrogen exchange effect during the 15N R2 relaxation time period. The H/H exchange rate is QkHX and the H/D exchange rate is (1-Q)kHX when the H2O content in the sample solvent is Q and the D2O content is (1-Q). In H/H exchange, the product NH′ is distinguished from the reactant NH.
To find kHX, the CLEANEX (Hwang et al. 1998) experiments were performed at pH 7.4 and pH 6.2 and measured kHX rates of αSyn were plotted in Fig. 3. The kHX values at pH 7.4 are significantly different from the values at pH 6.2 and also show variations across the protein sequence. Compared to the apparent R2CPMG values (Fig. 1) obtained from the single exponential decay, the difference in overall kHX values between pH 7.4 and pH 6.2 were much greater than the difference in apparent R2CPMG values indicating that the R2CPMG experiment suppresses a significant amount of the HX effect despite the fact that the HX effect is still observable on the apparent R2CPMG values.
Fig. 3.
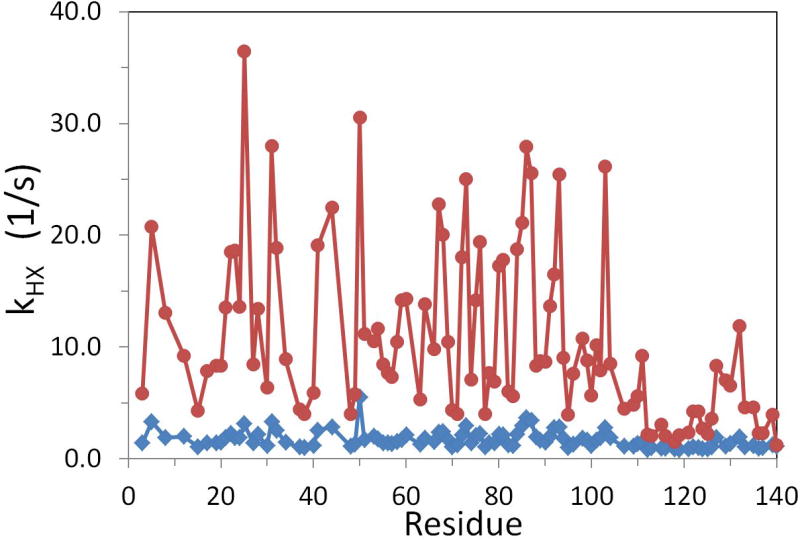
pH-dependent kHX values. kHX values of αSyn were measured by CLEANEX method at pH 7.4 (red circles) and pH 6.2 (blue rhombi) at 15 °C. The large increase of kHX values at pH 7.4 is prominent by increasing pH.
To ensure the steady state of amide and water proton magnetization, the R2CPMG pulse sequence with the proton SS pulse (Fig. 2) was used and R2CPMG relaxation data were acquired again at pH 7.4 and pH 6.2. With Q = 0.9, fanti = 0.011, and known kHX values from Fig. 3, multi-exponential fits to the R2CPMG relaxation data were performed with equation 5. The optimized R2CPMG values at pH 7.4 and pH 6.2 were plotted in Fig. 4. The large discrepancies in the apparent R2CPMG values of αSyn shown in Fig. 1 were eliminated by including the HX effect in the data analysis. R2CPMG values at pH 7.4 reported in Fig. 1 were corrected more than pH 6.2, R2CPMG values since kHX values at pH 7.4 were generally much greater than these values at pH 6.2. Average 15N R2CPMG values at pH 7.4 and pH 6.2 in Fig. 4 were 3.27 ± 0.46 Hz and 3.13 ± 0.44 Hz and the difference of average R2CPMG values between two pH’s was reduced to 0.14 Hz comparing to the difference of 0.67 in Fig. 1.
Fig. 4.
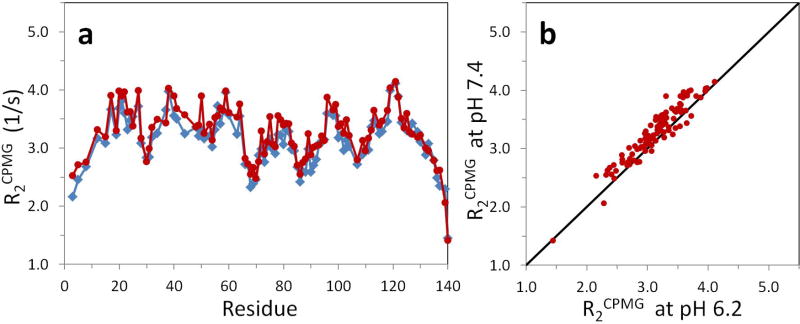
(a) Corrected R2CPMG values at pH 7.4 and 6.2. R2CPMG values at pH 7.4 (red circles) and pH 6.2 (blue rhombi) were obtained with the multi-exponential decay function (equation 5) using Q = 0.9, fanti = 0.011, and kHX values from Fig. 3. The relaxation data were acquired with the SS pulse. The large differences seen in Fig. 1 were corrected. (b) Correlation between R2CPMG values at pH 7.4 and pH 6.2.
To demonstrate how the HX impacts on the R2CPMG relaxation data, examples of the relaxation data with (filled squares) and without (filled circles) the proton SS pulse were plotted in Fig. 5 for residue G25 under fast HX (kHX = 36.3 Hz) at pH 7.4. Regardless of the SS pulse, multi-exponential fittings (solid lines) resulting from equation 5 improve the goodness of fit comparing to single exponential fittings (dotted line) in the presence of fast HX. Between multi-exponential fits of relaxation data with and without the proton SS pulse, it may be hard to judge whether the resulted R2CPMG values are correct or not by comparing the goodness of fits. However, R2CPMG values from the use of the SS pulse are smaller than those resulting from no SS pulse and represent the R2CPMG values without HX effect. The overall good agreement of the R2CPMG values between pH 7.4 and pH 6.2 (Fig. 4) can be achieved by acquiring the relaxation data with the proton SS pulse and analyzing the data with equation 5. Therefore, in the presence of observable HX, the R2CPMG relaxation data are affected by HX and these corrections are favorable.
Fig. 5.
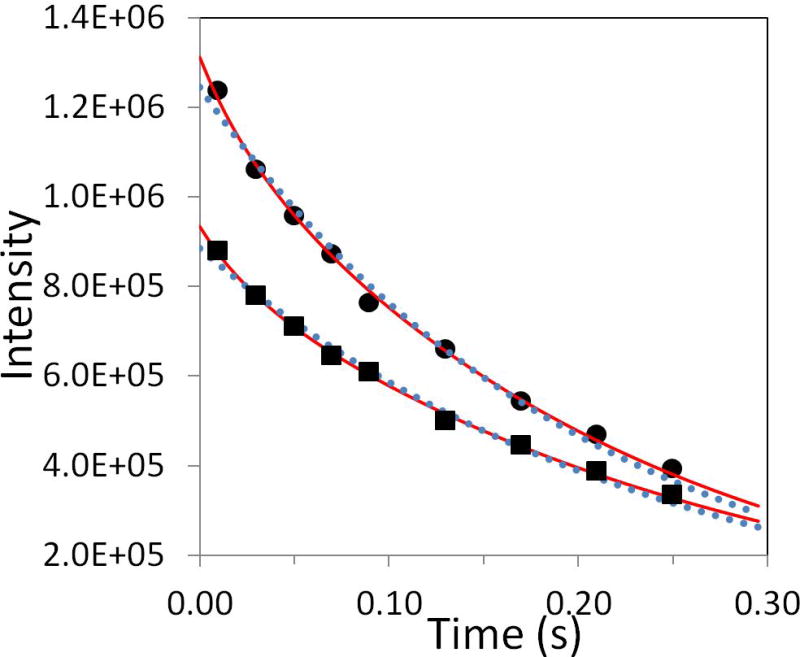
Examples of data fitting for residue G25. Experimental R2CPMG relaxation data for residue G25 at pH 7.4 were obtained with and without the proton SS pulse. The data (filled circles) without the proton SS pulse were fit with a single exponential (dotted line, I0 = 1.25×106, R2CPMG = 4.90 Hz, and R2 = 99.1 %) and a multi-exponential (equation 5) (solid line, I0 = 1.31×106, Q = 0.9, fanti = 0.011, kHX = 36.3 Hz, R2CPMG = 4.13 Hz, and R2 = 99.8 %), where R2 is the coefficient of determination for the goodness of fit. The data (filled squares) with the proton SS pulse were also fit with a single exponential (dotted line, I0 = 8.85×105, R2CPMG = 4.11 Hz, and R2 = 99.2 %) and a multi-exponential (equation 5) (solid line, I0 = 9.35×105, Q = 0.9, fanti = 0.011, kHX = 36.3 Hz, R2CPMG = 3.37 Hz, and R2 = 99.8 %).
In the presence of fast HX, it seems difficult to eliminate the HX effect in the R2CPMG experiment and also hard to identify the HX effect in the data analysis as shown in Fig. 5. Using a protein sample prepared in 100% H2O (Iwahara et al. 2007) will suppress the HX effect on R2CPMG data but incomplete suppression is still expected due to no control of the water proton magnetization by the steady state pulse and the anti-phase populations. Without correcting the HX effect, the elevated apparent R2CPMG values could be mis-interpreted as a result of other contributions such as conformational exchange or protein aggregation. In this context, conformational exchange of αSyn proposed at 37 °C may be due to HX effect on R2CPMG at these high temperatures (McNulty et al. 2006). When 1H-15N HSQC signal intensities are vulnerable by removing the water flip-back pulse and dephasing the water magnetization (Chen and Tjandra 2011), fast HX may play a role to attenuate the the signal intensity because of saturated water magnetization. In that situation, the HX effect needs to be under control in the acquisition scheme and in the data analysis as proposed here.
To minimize the HX effect on the R2CPMG experiment, relaxation rates need to be obtained by working under conditions for which fast HX does not exist such as low temperatures and low pH’s. In Supporting Information, dramatic intensity changes of HSQC spectra by HX effect were demonstrated by acquiring spectra at (pH 7.4, 35 °C), (pH 7.4 15 °C), (pH 6.1, 35 °C), and (pH 6.1, 15 °C). When sample conditions for removing fast HX cannot be obtained, we have shown that the 15N R2CPMG pulse sequence needs to contain SS pulses in order to remove water saturation effects and that quantitative corrections of the 15N relaxation rates for fast HX need to be performed. When the experiment is performed on a protein sample in mixed H2O/D2O solvent, use of the multi-exponential fit to the 15N R2CPMG relaxation data by equation 5 is a necessary to extract 15N R2CPMG values without HX effect. Other approaches that may simplify the data analysis include running the experiment in 100%H2O as practiced by Clore’s group (Iwahara et al. 2007). This removes the H/D exchange effect and the multi-exponential fit by equation 5 is changed to the single exponential fit with the rate constant of (fanti · kHX + R2CPMG). The required HX rates in data analysis can be measured independently from R1zz (Skrynnikov and Ernst 1999), MEXICO (Gemmecker et al. 1993), CLEANEX (Hwang et al. 1998), or 15NH/D-SOLEXSY (Chevelkov et al. 2010) experiments. As an alternative to R2CPMG, the 15N R1ρ experiment has been also used for R2 measurement after correction of off-resonance effects (Korzhnev et al. 2002; Palmer III et al. 2001). If the anti-phase population is negligible (fanti ≅ 0) in the 15N R1ρ experiment by applying a strong spin-lock on Nx, then equation 5 becomes the single exponential fit with the rate constant of R1ρ with the use of 100% H2O. A noticeable merit of this approach is no requirement of measuring kHX values. However, it is important that the SS pulses are used for any variation of experiments to assure the steady state HX equilibrium during the inter-scan recycle delay.
Although we demonstrated the HX effect on the backbone 15N R2CPMG relaxation data applied to the IDP αSyn, the HX effect may be present in other protein samples and various NMR experiments. In addition to IDPs, globular proteins with unstructured elements such as large loop regions, or partially folded proteins that have a large number of amides exposed to the solvent may have increased apparent 15N R2CPMG values due to fast HX. Other NMR experiments that have long evolution times to measure relaxation and diffusion should be potentially considered for modulation by fast HX effects as well (Dempsey 2001; Zhang et al. 1995). Like 15N R2CPMG experiments, NMR experiments that use the indirect detection scheme also need to be considered with caution although heteronuclear NMR properties other than 1H are observed.
Supplementary Material
Acknowledgments
K-PW was supported by an NIH Interdisciplinary Research Workforce Fellowship (5R90DK071502). This work has been supported by NIH grants (GM087012 and GM45302) and NSF grants (DBI-0403062 and DBI-0320746) to JB.
References
- Bertini I, Carrano CJ, Luchinat C, Piccioli M, Poggi L. A 15N NMR mobility study on the dicalcium P43M calbindin D9k and its mono-La3+-substituted form. Biochemistry. 2002;41:5104–5111. doi: 10.1021/bi015945n. [DOI] [PubMed] [Google Scholar]
- Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO. Structural characterization of the intrinsically unfolded protein beta-synuclein, a natural negative regulator of alpha-synuclein aggregation. J Mol Biol. 2007;372:708–722. doi: 10.1016/j.jmb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Buevich AV, Baum J. Dynamics of unfolded proteins: incorporation of distributions of correlation times in the model free analysis of NMR relaxation data. J Am Chem Soc. 1999;121:8671–8672. [Google Scholar]
- Buevich AV, Shinde UP, Inouye M, Baum J. Backbone dynamics of the natively unfolded pro-peptide of subtilisin by heteronuclear NMR relaxation studies. J Biomol NMR. 2001;20:233–249. doi: 10.1023/a:1011243116136. [DOI] [PubMed] [Google Scholar]
- Bussell R, Jr, Eliezer D. Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol Chem. 2001;276:45996–46003. doi: 10.1074/jbc.M106777200. [DOI] [PubMed] [Google Scholar]
- Chen J, Tjandra N. Water proton spin saturation affects measured protein backbone 15N spin relaxation rates. J Magn Reson. 2011;213:151–157. doi: 10.1016/j.jmr.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevelkov V, Xue Y, Rao DK, Forman-Kay JD, Skrynnikov NR. 15NH/D-SOLEXSY experiment for accurate measurement of amide solvent exchange rates: application to denatured drkN SH3. J Biomol NMR. 2010;46:227–244. doi: 10.1007/s10858-010-9398-8. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Croke RL, Sallum CO, Watson E, Watt ED, Alexandrescu AT. Hydrogen exchange of monomeric alpha-synuclein shows unfolded structure persists at physiological temperature and is independent of molecular crowding in Escherichia coli. Protein Sci. 2008;17:1434–1445. doi: 10.1110/ps.033803.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dempsey CE. Hydrogen exchange in peptides and proteins using NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2001;39:135–170. [Google Scholar]
- Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer D, Kutluay E, Bussell R, Jr, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandham R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Gemmecker G, Jahnke W, Kessler H. Measurement of fast proton exchange rates in isotopically labeled compounds. J Am Chem Soc. 1993;115:11620–11621. [Google Scholar]
- Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci. 2001;2:492–501. doi: 10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- Griesinger C, Ernst RR. Cross relaxation in time-dependent nuclear spin systems: Invariant trajectory approach. Chem Phys Lett. 1988;152:239–247. [Google Scholar]
- Hansen DF, Led JJ. Implications of using approximate Bloch–McConnell equations in NMR analyses of chemically exchanging systems: application to the electron self-exchange of plastocyanin. J Magn Reson. 2003;163:215–227. doi: 10.1016/s1090-7807(03)00062-4. [DOI] [PubMed] [Google Scholar]
- Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Jung YS, Clore GM. Heteronuclear NMR spectroscopy for lysine NH3 groups in proteins: unique effect of water exchange on 15N transverse relaxation. J Am Chem Soc. 2007;129:2971–2980. doi: 10.1021/ja0683436. [DOI] [PubMed] [Google Scholar]
- Kateb F, Pelupessy P, Bodenhausen G. Measuring fast hydrogen exchange rates by NMR spectroscopy. J Magn Reson. 2007;184:108–113. doi: 10.1016/j.jmr.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE. An NMR Experiment for the Accurate Measurement of Heteronuclear Spin-Lock Relaxation Rates. J Am Chem Soc. 2002;124:10743–10753. doi: 10.1021/ja0204776. [DOI] [PubMed] [Google Scholar]
- Mandel AM, Akke M, Palmer AG., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- McConnell HM. Reaction rates by neclear magnetic resonance. J Chem Phys. 1958;28:430–431. [Google Scholar]
- McNulty BC, Tripathy A, Young GB, Charlton LM, Orans J, Pielak GJ. Temperature-induced reversible conformational change in the first 100 residues of alpha-synuclein. Protein Sci. 2006;15:602–608. doi: 10.1110/ps.051867106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Mittermaier AK, Kay LK. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Palmer AG., III NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Palmer AG, III, Kroenke CD, Loria JP. Nuclear Magnetic Resonance Methods for Quantifying Microsecond-to-Millisecond Motions in Biological Macromolecules. Method Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Palmer AG, III, Skelton NJ, Chazin WJ, Wright PE, Rance M. Suppression of the effects of cross-correlation between dipolar and anisotropic chemical shift relaxation mechanisms in the measurement of spin-spin relaxation rates. Mol Phys. 1992;75:699–711. [Google Scholar]
- Skrynnikov NR, Ernst RR. Detection of intermolecular chemical exchange through decorrelation of two-spin order. J Magn Reson. 1999;137:276–280. doi: 10.1006/jmre.1998.1666. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Quantitative NMR studies of high molecular weight proteins: application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G. J Mol Biol. 2003;327:1121–1133. doi: 10.1016/s0022-2836(03)00238-9. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Wang C, Palmer AG., 3rd Solution NMR methods for quantitative identification of chemical exchange in 15N-labeled proteins. Magn Reson Chem. 2003;41:866–876. [Google Scholar]
- Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K-P, Kim S, Fela DA, Baum J. Characterization of conformational and dynamic properties of natively unfolded human and mouse alpha-synuclein ensembles by NMR: implication for aggregation. J Mol Biol. 2008;378:1104–1115. doi: 10.1016/j.jmb.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K-P, Weinstock DS, Narayanan C, Levy RM, Baum J. Structural Reorganization of alpha-Synuclein at Low pH Observed by NMR and REMD Simulations. J Mol Biol. 2009;391:784–796. doi: 10.1016/j.jmb.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, Paterson Y, Roder H. Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein Sci. 1995;4:804–814. doi: 10.1002/pro.5560040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


