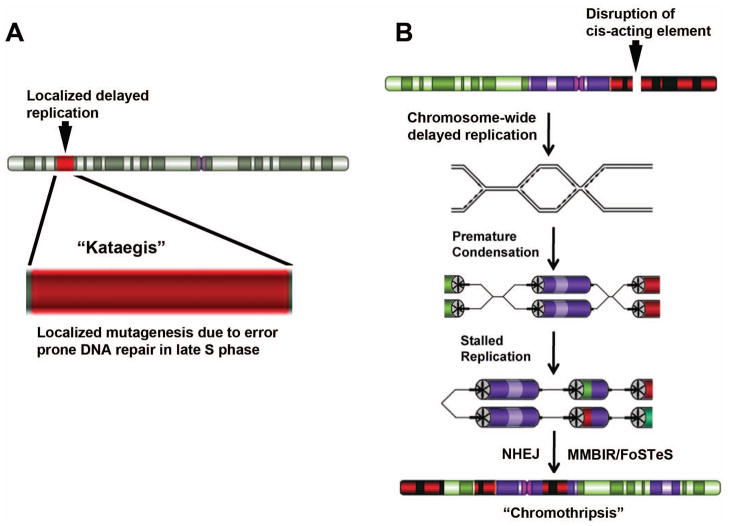
Figure 2.
Models for localized genomic instability in cancer cells. A) Aberrant late replication model for kataegis. A localized region of a chromosome has acquired abnormally late replication (red) either as a result of chromosome rearrangement or as a result of a localized shift in the replication timing program [16]. Increased mutagenesis is induced in the late replicating region due to error prone repair mechanisms functioning during late replication. B) Aberrant late replication model for Chromothripsis. Disruption of discrete cis-acting loci result in a chromosome-wide delay in replication timing. Mitotic chromosome condensation initiates on the delayed chromosome prior to completion of DNA synthesis resulting in premature chromosome condensation, stalled replication forks, and rearrangement of the affected chromosomes via non-homologous end joining (NHEJ), microhomology mediated break induced replication (MMBIR) and fork stalling and template switching (FoSTeS) mechanisms. The resulting chromosome contains numerous structural alterations (translocations, deletions, inversions, and duplications).