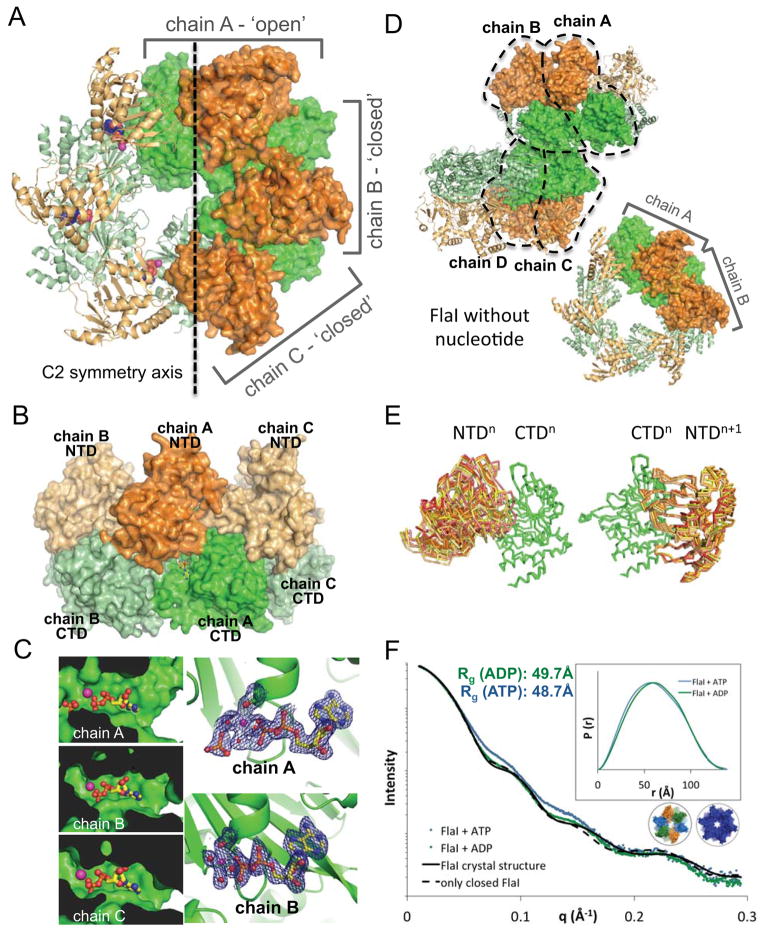
Figure 2. FlaI crystal structures show different subunit conformations.
(A) Crystal structure of the Walker B mutant FlaIE336A with NTDs (orange) and CTDs (green). A C2 symmetry axis divides the hexameric ring into 2 halves, three chains form the asymmetric unit (surface representation): chain A adopts a much more open conformation than chains B and C.
(B) The FlaI ring forms a crown-like structure. Side view of the FlaIE336A hexameric ring shows the crown-like structure with the CTDs forming the base, the NTDs the points, with nucleotide shown as yellow sticks.
(C) The three active sites are not identical. The active site in chain A is more open and solvent accessible than in chains B and C. Electron density (2|Fo|-|Fc| map at 1.5σ) for released phosphate is seen in chain A but not for chains B and C, which have similar electron densities (chain B shown as an example).
(D) Apo-FlaI without nucleotide also crystallized as a hexameric ring. Two subunits from two different hexameric rings form the asymmetric unit (surface representation), resulting in four unique subunit conformations. Colored as in A.
(E) Comparison of the seven observed FlaI conformations reveals FlaI building blocks (three from ADP-FlaI and four from apo-FlaI). Left, alignment of the CTDs shows many different conformations of the corresponding NTDs. Right, Building blocks of a CTDn with NTDn+1 superimpose very well for all seven conformations.
(F) ATP-bound FlaI is more compact than ADP-FlaI. SAXS data of FlaI with ATP (blue) and ADP (green) shows a more compact structure for ATP-FlaI. Theoretical scattering curves for the crystal structure (black line) and an all closed model (dashed black line) are shown. Inset: P(r) distribution of the same data. See also Figure S2.