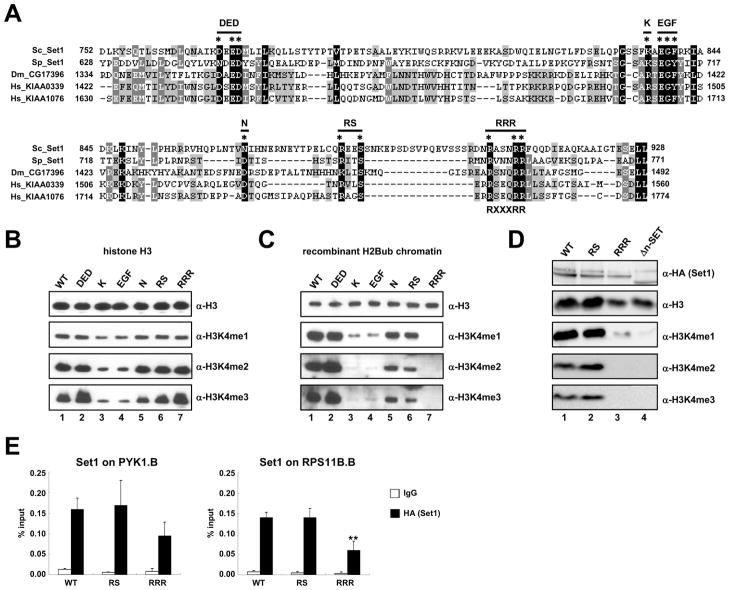
Figure 5. Identification of Amino Acid Residues within the n-SET Domain Responsible for H2B Ubiquitylation-Dependent H3K4 Methylation Activity of the Yeast Set1 Complex.
(A) ClustalW2 multiple sequence alignments of n-SET domains from Set1 family members: Saccharomyces cerevisiae (Sc_Set1, GeneBank Accession number: NP_011987), Schizosaccharomyces pombe (Sp_Set1, NP_587812), Drosophila melanogaster (Dm_CG17396, NP_001015221) and Homo sapiens (Hs_KIAA0339, BAA20797; Hs_KIAA1076, Q9UPS6). Encoded amino acid numbers are indicated. Conserved amino acids that were changed to alanine are marked by asterisks, and labels for derived Set1 mutants are indicated at the top. The consensus for the RXXXRR motif is indicated at the bottom.
(B and C) In vitro HMT assays with purified ySet1Cs bearing the indicated Set1 mutations and free histone H3 (B) or H2Bub-chromatin templates (C).
(D) Immunoblot analyses with indicated antibodies of whole cell extracts from yeast strains that carry chromosomal genes expressing the indicated HA-tagged WT or mutant Set1 proteins.
(E) ChIP analyses with anti-HA antibody to determine chromatin association of HA-tagged WT, RS and RRR Set1 proteins at 5′-transcribed regions (marked ‘B’ in Figure 4D) of PYK1 (left) and RPS11B (right) genes. The significance of the differences in the ChIP signals was evaluated using the Student’s t-test (** denotes p-value < 0.01).
See also Figure S5.