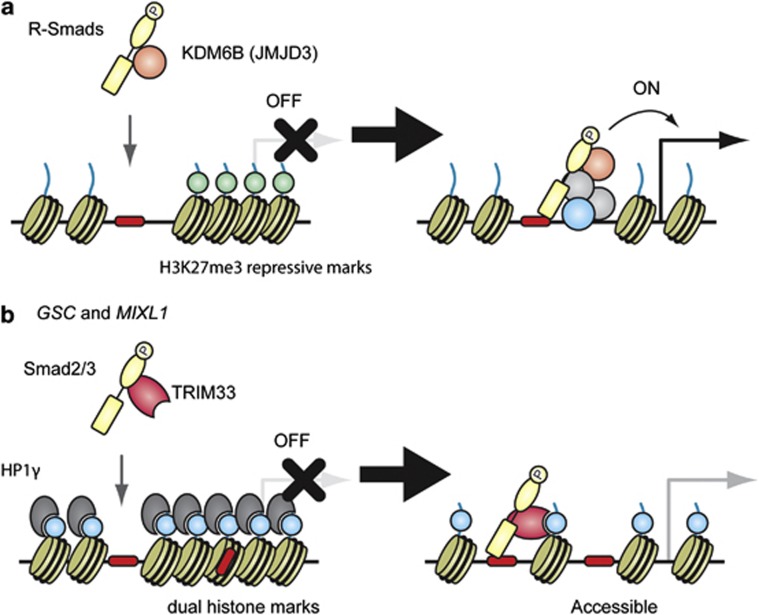
Figure 3.
Smad proteins and histone modification marks. Smad proteins have been reported to induce local chromatin remodeling and modification at their binding sites. Several models are described in ES cells, where early developmental genes are poised and ready to be activated in response to extracellular signals, such as nodal. (a) R-Smads physically interact with a histone demethylase, KDM6B (also known as JMJD3), and recruit it to their target sites, followed by the loss of the H3K27me3 repressive mark (light green). (b) Xi et al.70 reported that nodal signaling triggered TRIM33–Smad2/3 complex formation. The TRIM33–Smad2/3 complex recognizes and binds to H3K9me3-K18ac dual histone marks (light blue) and displaces the chromatin-compacting factor HP1γ (heterochromatin protein 1γ) in the GSC and MIXL1 promoters, resulting in the remodeling of the local chromatin structure to make Smad-binding region(s) (red) accessible. A full colour version of this figure is available at the Oncogene journal online.