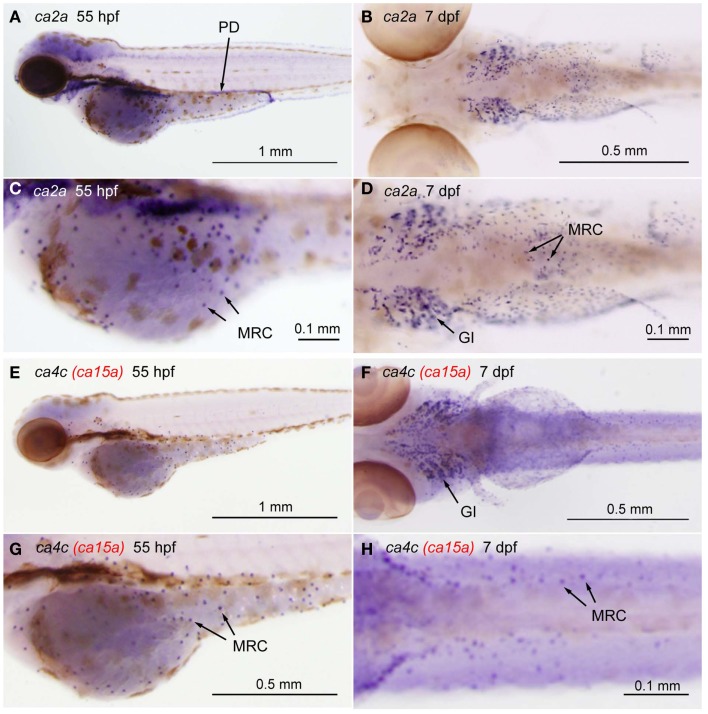
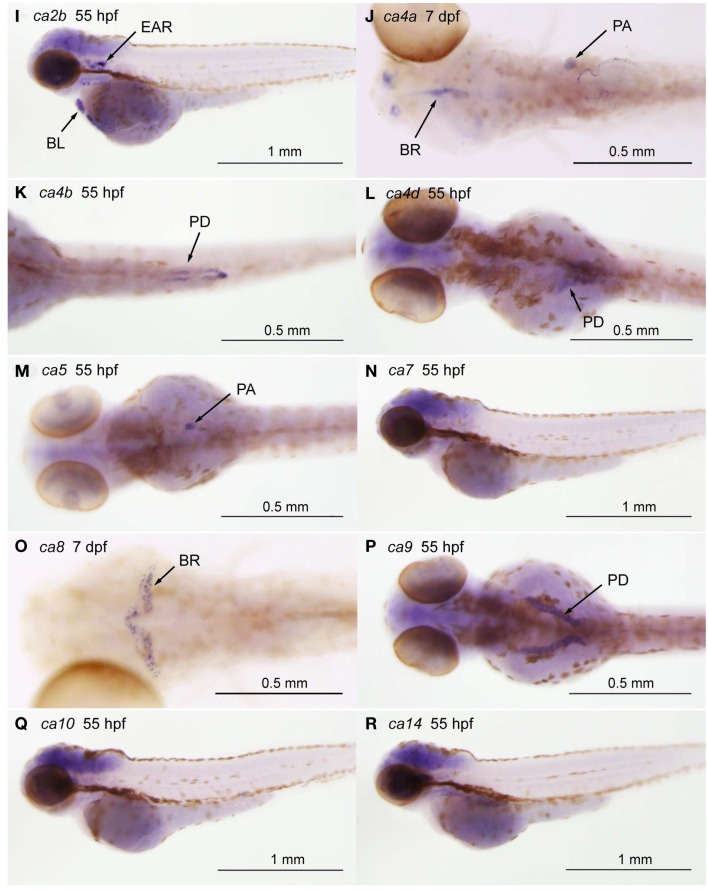
Figure A1.
Tissue-specific expression of CA isoforms in zebrafish larvae. Expression of ca genes was analyzed in 55-hpf and 7-dpf larvae by whole mount in situ hybridization with cRNA probes against ca2a [(A,C), lateral views and (B,D), ventral views], ca2b [(I), lateral view], ca4a [(J), dorsal view], ca4b [(K), ventral view], ca4c or ca15a [(C,E), lateral views and (D,F), ventral views], and ca4d [(L), dorsal view], ca5 [(M), dorsal view], ca7 [(N), lateral view], ca8 [(O), dorsal view], ca9 [(P), dorsal view], ca10 [(Q), lateral view], and ca14 [(R), lateral view]. (C,D,G,H) Magnified views of (A,B,E,F), respectively. Tissue- or cell-specific expression of individual ca genes was indicated with arrows in each panel. Abbreviations are gill (GI), mitochondria-rich cell (MRC), blood cell (BL), brain (BR), pronephric ducts (PD), pancreas (PA). We note that broad staining in the brain was non-specific background as it was observed with sense cRNA probes. Four CA4 isoforms predicted from zebrafish database showed different tissue-specific distribution. ca4a, most homologus to mammalian CA IV, is expressed in the pancreas and expected to be involved in the regulation of the luminal pH of the pancreatic duct system (Fujikawa-Adachi et al., 1999). The other two isoforms ca4b and ca4d are expressed respectively in the posterior part and anterior part of the renal pronephric ducts. These expressions reminds that localization of mammalian membrane-bound CA in the proximal tubule cells and the intercalated cells of the collecting ducts, which is suggested to contribute to reabsorption of and regulation of acid-base balance of urine (Dobyan and Bulger, 1982). Its location suggests a role in regenerating CO2, a substrate for CA II. For ca4c (i.e., ca15a), see the text.