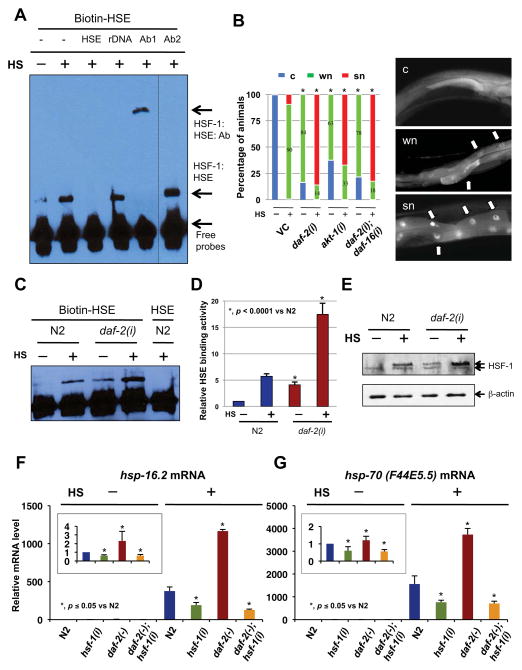
Figure 1. Inactivation of daf-2 positively regulates HSF-1 activity and heat-shock response.
(A) DNA-binding activity of C. elegans HSF-1 in response to 90 min of heat-shock at 37°C (HS), measured by electrophoretic mobility shift assay. Nuclear extracts were incubated with biotin-labeled HSE probes (Biotin-HSE). The specificity of the DNA binding was determined by competition with 200x unlabeled HSE probes (HSE), randomly synthesized DNA (rDNA), anti-HSF-1 antibodies (Ab1), or anti-GFP antibodies (Ab2) before being applied to gel eletrophoresis. (B) Nuclear accumulation of HSF-1 in response to IIS inactivation. EQ73 animals (hsf-1::gfp) grown on vector control (VC) or different RNAi bacteria were unstressed or heat-shocked for 30 min (HS) before being classified into three groups according to the nuclear/cytosolic (n/c) ratio of GFP intensity in the intestinal cells (right panels). “c”, “wn”, and “sn” are animals with n/c ratio < 1.2, 1.2~2.0, and >2.0, respectively. The mean of three independent experiments were pooled and shown (left panel). *, p < 0.0001 vs VC under same conditions (chi2-test). n ≥ 300. (C–D) The DNA-binding activity of HSF-1 in daf-2(e1370) mutants in response to 90 min of heat-shock (HS). The result of a representative experiment is shown in (C). The mean ± SD of three independent experiments (mean ± SD), normalized to the control (N2 with unlabeled HSE), is presented in (D). (E) N2 or daf-2(RNAi) animals were unstressed or heat-shocked for 90 min (HS). Worm whole cell extracts (WCE) of these animals were subjected to immunoblotting analysis using anti-HSF-1 (top) or anti-β-actin (bottom) antibodies. Detail quantification in Fig. S1D. (F–G) Relative abundance of (F) hsp-16.2 and (G) hsp-70 (F44E5.5) mRNA in wild types (N2) or daf-2(e1370) mutants fed with control or hsf-1 RNAi bacteria. The inset shows the mRNA level under unstressed conditions (without 90 min HS). Data were combined from at least three experiments, and the mean ± SD of each treatment are shown.