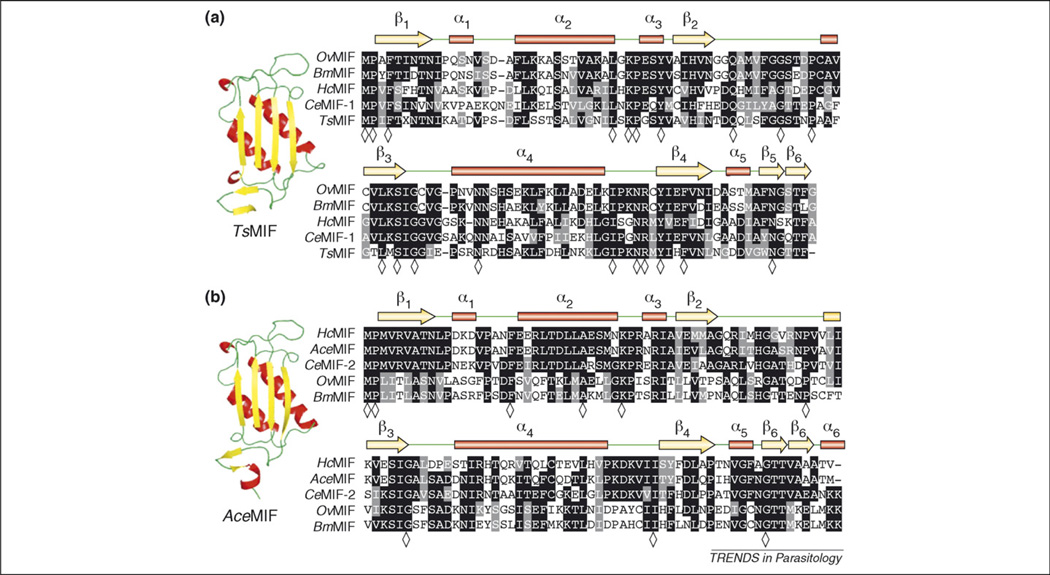
Figure 2.
Multiple sequence alignments of nematode MIFs. (a) Type 1 MIFs, including prototype CeMIF-1, with corresponding secondary amino acid structure assignments listed above alignment including α-helices represented as red bars, β-strands as yellow arrows and random coils in green. Representative ribbon backbone diagram of type 1 MIF member Trichinella spiralis MIF (TsMIF). (b) Type 2 MIFs, including prototype CeMIF-2, with corresponding secondary amino acid structure assignments as described above. Representative ribbon backbone diagram of type-2 MIF member Ancylostoma ceylanicum MIF (AceMIF). Sequence alignments were generated with Clustalw and BoxShade programs. Identical residues are shaded in black and conserved residues are shaded in gray; residues identical in alignments of all type 1 (21 sequences) or type 2 (14 sequences) MIFs from Table 1 are indicated in the consensus line with a diamond (◇).