Abstract
The Pittsburgh Center of Excellence in Public Health Informatics has developed a probabilistic, decision-theoretic system for disease surveillance and control for use in Allegheny County, PA and later in Tarrant County, TX. This paper describes the software components of the system and its knowledge bases. The paper uses influenza surveillance to illustrate how the software components transform data collected by the healthcare system into population level analyses and decision analyses of potential outbreak-control measures.
1. Introduction
The Center for Advanced Study of Informatics in Public Health (CASIPH) is developing and integrating software to create a probabilistic, decision-theoretic system for disease surveillance and control and is translating this system into practice at the Allegheny County Health Department (ACHD).
This work represents a new paradigm for disease surveillance, based on probability and decision theory. The approach integrates Bayesian diagnosis of individual patients with Bayesian “diagnosis” of a population. It is capable of estimating the current incidence of influenza in a population from data in electronic medical records (EMRs). It is also able to estimate the set of parameters required to initialize a SEIR epidemic model. It is thus possible to initialize an epidemic model so as to match the current disease status of a population. The epidemic model is then used in a decision model of available control measures. This unique capability will enable decision makers to use epidemic models more effectively when selecting control measures for influenza and other outbreaks.
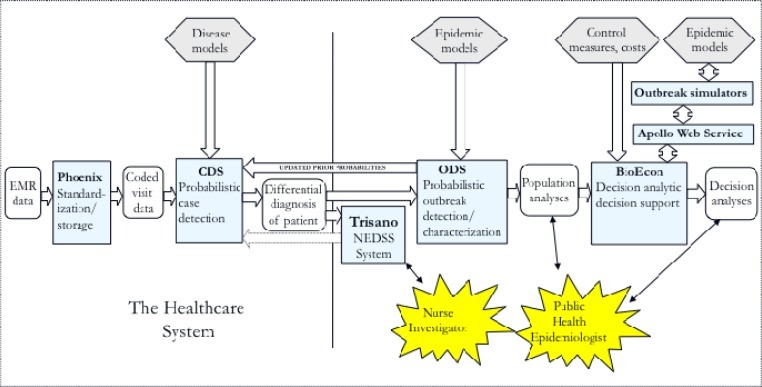
Figure 1 shows the software components (blue rectangles) of the probabilistic, decision-theoretic system and how they combine to form an end-to-end disease surveillance system. This paper describes each software component, how information flows in the system, and the level of integration of the components that we have achieved to date.
Figure 1.
Information flow in the probabilistic, decision-theoretic disease surveillance system.
Legend: light blue rectangles, software components: CDS, Case Detection System; ODS, Outbreak Detection and characterization System; hexagons, knowledge bases; rounded rectangles, data and analyses; thin arrows, user interfaces; large arrows indicate that the components exchange data electronically in the direction indicated; dotted arrow indicates a future connection sending case data collected by nurse investigators from Trisano® to CDS.
2. Software Components
This section describes the software components depicted in Figure 1.
2.1. Phoenix
The role of Phoenix is to receive patient data from an EMR and respond to queries from the case detection system for patient data in standard form. Phoenix receives EMR data as HL7 messages. It then parses the messages, standardizes the extracted information, and stores it in a database.
Phoenix comprises:
A database: The database uses multiple entity-attribute-value tables, a representation that we found necessary to meet the real-time processing requirement of the project. Our initial implementation of the database used OpenMRS, which we extended with our own relational database schema. However, OpenMRS was unable to keep up with the volume of patient data from the UPMC health system even after we modified it to run on the Oracle database management system to take advantage of Oracle’s scalability and database optimizations. Even with Oracle’s optimizations and scalability, a relational table design was too slow, necessitating the entity-attribute-value representation that we use currently.
HL7 parsers, for microbiology, chemistry, dictation, ADT, and radiology HL-7 feeds
A data viewer, which gives a clinical episode (e.g., emergency room visit) view of the data for internal development purposes and serves as a prototype for future end-user interface for a health department.
A data standardization program, which converts local UPMC Health System terminology to standard codes (UMLS, SNOMED-CT, LOINC).
Phoenix is not a primary focus of our work; it can be understood as necessary scaffolding that we had to construct to access EMR data and allow for the use of standard terminology in the rest of the components. We intend to discard it when EMRs are capable of providing Phoenix’s functionality to public health applications.
2.2. Bayesian Case Detection System (CDS)
The role of CDS is to infer a patient’s medical diagnosis from data in an EMR. In particular, CDS computes a probabilistic differential diagnosis for each disease in a set of monitored diseases for every patient visit to a monitored facility. A probabilistic differential diagnosis is a list of diseases with their posterior probabilities, given the available data. At present, CDS processes all emergency department (ED) visits in monitored hospitals in Allegheny County, PA. It computes both a posterior probability of influenza and a likelihood, P(patient data|influenza) for each visit. It uses information extracted from ED dictations by a natural language processing algorithm as patient data for this inference. CDS also computes a posterior probability for the majority of notifiable diseases in Allegheny County from the laboratory test results of ED patients.
CDS uses Bayesian networks to compute a patient’s differential diagnosis. A Bayesian network is a directed graph in which the nodes represent variables and probabilities distributions for the variables, given its parents (indicated by the directed arcs in the network).A Bayesian network is a compact factorization of the joint probability distribution over the variables. A Bayesian inference algorithm can compute any marginal or conditional probability from this factorization. In particular, it can compute the probability that a patient has influenza, given that the patient has fever, but not cough.
The influenza Bayesian network in CDS comprises a node that represents the diagnosis influenza and a set of nodes that represent the symptoms, signs, and laboratory results that may contribute to a diagnosis of influenza. There is a single diagnosis node and 367 finding nodes in the influenza Bayesian network. The diagnosis node takes one of two values: true and false, where false means that the patient has an illness other than influenza. The finding nodes take one of three values: present, absent, and unknown, where unknown means that the natural language processing cannot determine a value for the finding from the patient data.
The notifiable-disease Bayesian networks represent only laboratory tests, at present. Collectively, the set of “lab-only” Bayesian networks functions as a probabilistic Electronic Laboratory Reporting (ELR) system, which has equivalent functionality to the existing ELR paradigm when its probabilistic thresholds for reporting are set close to 1.0. Ultimately, we intend for CDS to contain diagnostic models similar to the influenza Bayesian network for all notifiable conditions, syndromes of interest, and emerging diseases.
The CDS runs once per day, at present, although it can process ED visits in real time. In its current configuration, CDS obtains patient data from Phoenix for all patient ED visits that occurred in the previous 24 hours. CDS then computes both the disease likelihood and the posterior probability of influenza and for the monitored notifiable diseases for each patient visit. Only the disease likelihoods for influenza, P (findings | influenza), are sent to the Outbreak Detection and Characterization System (ODS) described in Section 2.4. The data sent to ODS for each visit also include the date of visit, home zip code, patient age in deciles, and patient gender, although they are not used by ODS at present.
CDS has been in production operation in Allegheny County since 2009. It sends a daily report by email to the health department. The report plots the CDS estimate of the number of influenza cases seen in the monitored EDs. To generate the estimated number of influenza cases for a day, CDS sums the posterior probabilities of every patient seen in the EDson that day to form an expected number of cases for the day.
Tsui et al. describe CDS in detail in an accompanying paper in this issue of OJPHI.
2.3. TriSano®
TriSano® Community Edition (CE) is open source “NEDSS” software used by the Utah Department of Health (http://www.trisano.com/).TriSano® is representative of case investigation software in use by other health departments. We are extending Trisano® to support bidirectional communication with CDS, as depicted in Figure 1.
In the CDS-to-Trisano® “reporting” direction, we have implemented the capability for CDS to transmit cases to TriSano® and for a TriSano® end-user to set a disease-specific reporting threshold, Td, for a given disease d, within that application. When P (d= present | patient data) >Td, the case appears in the TriSano® inbox.
In a future Trisano®-to-CDS connection, TriSano® will send case data recorded by nurse investigators to CDS so that CDS can recompute the patient’s probabilistic differential diagnosis and in turn update ODS. We also expect that the additional information and updated diagnosis may be of value in the clinical care of patients.
2.4. ODS
The function of the Bayesian Outbreak Detection and Characterization System (ODS) is to detect outbreaks and to estimate outbreak characteristics, such as infectious period, latent period, and Ro (reproductive rate).
ODS integrates tightly with CDS. As was stated earlier, for each patient who visits a monitored ED, CDS determines how likely that patient’s findings match each of a set of modeled outbreak diseases (e.g., influenza). Such a match is expressed as the probability of the findings given a disease, namely, patient disease likelihoods. ODS takes as input these likelihoods, plus the prior probability distribution over the various types of outbreak diseases being modeled. It then samples from distributions representing the input parameters to an epidemic model for a given disease type. When using a SEIR epidemic model of influenza, for example, ODS samples from distributions that are characteristic of influenza for the infectious period, latent period, Ro, initial number infected, and start date. It then derives the posterior probability of each sampled model.
More formally, let MPop denote an epidemic model of the entire population in a region that is being monitored for an outbreak of disease. In our current application, MPop is a SEIR model. We would like to infer a distribution over such models given evidence about patients who seek care at emergency departments in the region. Let MED denote the disease states of all the ED patients during the monitoring period. Let E designate the clinical evidence that is available about the ED patients during the monitoring period.
At a high level, ODS is based on the following equation, which is an instance of Bayes’ theorem:
The sum is taken over all possible disease states of all the ED patients being monitored. The number of terms in the sum is therefore very large; however, we are able to take advantage of some basic mathematical techniques, such as the application of the binomial distribution, to compute the sum efficiently. ODS approximates the integral in the equation by sampling MPop, which leads to the integral becoming a sum. The terms P(MED | MPop) and P(E | MED) represent key modeling components of ODS. The term P(MPop) represents a prior probability distribution over the parameters in a SEIR model of the population. The independence assumption in the equation is that P(E | MED, MPop) = P(E | MED), which expresses that in predicting ED patient data, knowledge of the disease status of the population at large is irrelevant, once we have knowledge of the disease status of the ED patients.
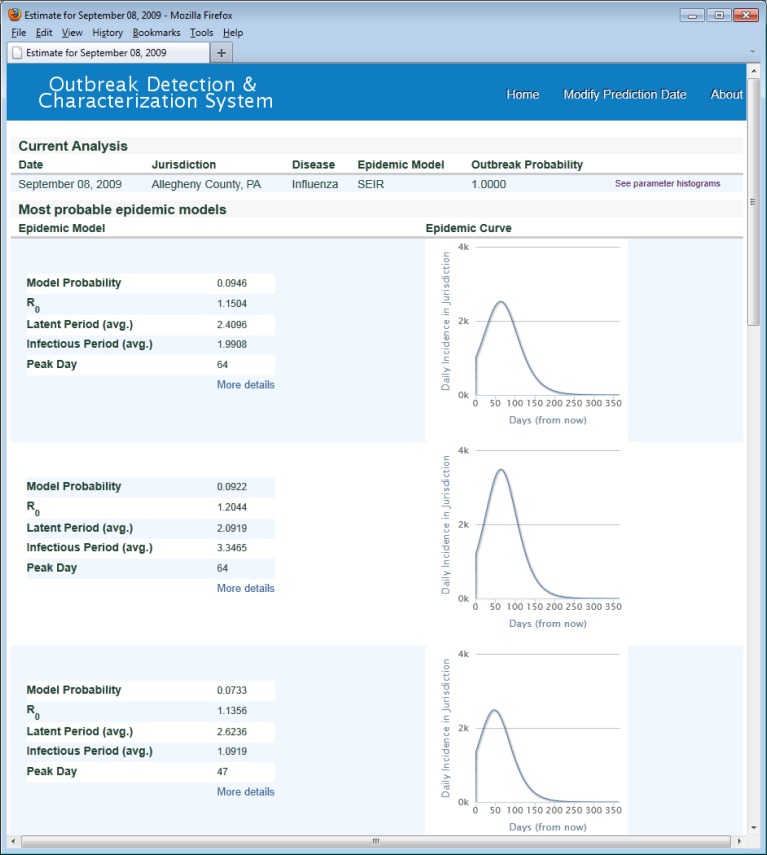
Figure 2 shows the most probable models computed by ODS using ED EMR data from the UPMC Health System in Allegheny County (AC) through September 7, 2009 (retrospective analysis). On September 7, 2009, the influenza surveillance data were beginning to show influenza activity in AC.
Figure 2:
ODS, most probable models.
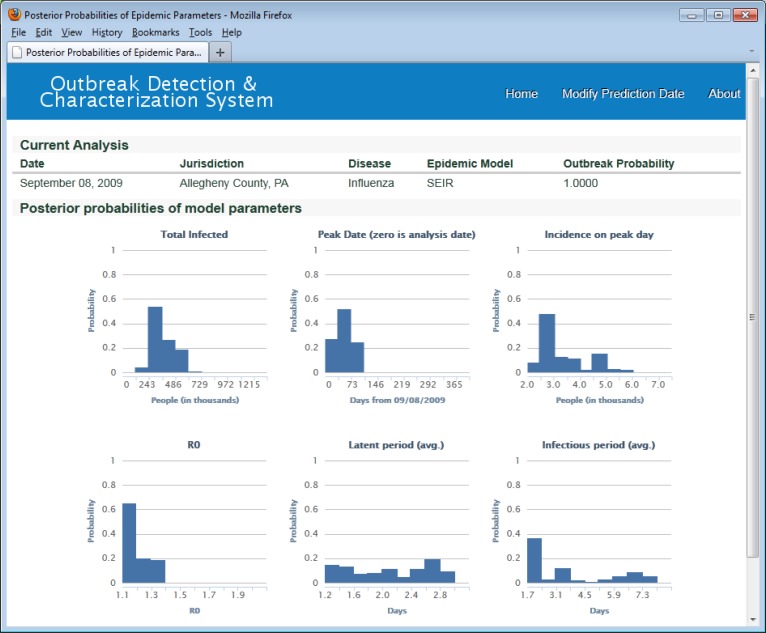
Figure 3 shows posterior distributions over three SEIR model parameters—Ro, infectious period, and latent period (bottom three graphs). These histograms show the parameters of the 269 most likely epidemic models on September 8, 2009. The set of 269 most likely epidemic models were those whose cumulative posterior probability summed to an arbitrary threshold p > 0.99995. Figure 3 also shows distributions for peak date, incidence on peak day, and total number infected for the set of 269 most likely epidemic models (top three graphs).
Figure 3.
Distributions of total infected, peak date, incidence on peak day, and posterior distributions for Ro, latent period, and infectious period. ODS used uniform prior probabillity distributions for SEIR model parameters in this analysis. For example, the prior distribution for Ro was uniform over the range 1.1 to 1.9.
For the H1N1 outbreak, the most probable ODS model predicted that the number of infected individuals in AC would peak on November 11, 2009. Based on laboratory measurements of influenza reported in (1), the H1N1 epidemic is believed to have peaked in AC between October 24th and November 7th. Thus, based on ED patient data that was available about seven weeks prior to the actual peak, the most probable model predicted the actual peak quite accurately. Based also on laboratory measurements, by approximately November 23, 2009 the percentage of people in AC who had been infected with H1N1 was estimated to be about 21% (1). Again, using data only up through September 8, 2009, the most probable model predicted that about 19% of the AC population would be infected by November 23rd, which is close to the actual percentage.
This case study, which involved real data and an actual influenza outbreak, provides support that the basic approach outlined above is promising. However, it is only one influenza outbreak, and thus, additional study of the approach is clearly needed.
The above approach could be applied to other diseases and it can be generalized to use other types of epidemic models, including segmented compartment models and agent-based models.
ODS also provides CDS with dynamically updated ED priors for influenza. These priors can be combined with the likelihoods computed by CDS to obtain a posterior probability of influenza for each ED patient, based on a real-time estimate of influenza prevalence in the ED; in turn, these posteriors can be used to support clinical decision making for individual patients (e.g., decisions about testing and treating for influenza).
This screen shows the three most probable SEIR influenza models given the likelihoods of influenza from CDS for all Allegheny County patients seen in monitored UPMC EDs from May through September 7, 2009. On the daily incidence curves, day zero is September 8, 2009.
Prior work using Bayesian algorithms for disease surveillance has had an emphasis on detection rather than characterization. Examples of temporal methods include (28), who extended Kulldorff’s spatial scan statistic to produce posterior probabilities of influenza in geographical sub-regions. A multivariate generalization was developed in (14). Spatio-temporal approaches include the WSARE 3.0 algorithm (15), the PANDA algorithm for detecting anthrax outbreaks (16), the PCTS algorithm for detecting outbreaks of all CDC Category A diseases that are of special concern for biosurveillance (17), and a Bayesian hierarchical model to detect anomalously high levels of influenza (18). In previous research, we developed Bayesian algorithms (16, 17) that employed a data likelihood approach, similar to the method we describe here. However, they were based only on chief complaints as evidence.
Our approach to outbreak detection and characterization (OD&C) has important features not present in previous work. First, instead of analyzing counts of data to estimate an epidemic curve (19, 20), we use a flexible and more general approach that models probabilistically the available evidence, such as the rich set of patient findings in ED reports. The approach reflects the intrinsic synergy between individual patient diagnosis and population OD&C. In particular, OD&C is derived based on past probabilistic patient diagnoses. In turn, the diagnosis of a newly arriving patient is based on prior probabilities that are derived from probabilistic inference over current OD&C models. To our knowledge, no prior research (either Bayesian or non-Bayesian) has taken such an integrated approach to patient diagnosis and population OD&C.
Second, our approach represents a general Bayesian framework for modeling OD&C. It can be applied with many different types of disease outbreak models including SEIR (Susceptible, Exposed, Infectious, and Recovered) model (21), agent-based, and outdoor-substance-release (OSR) models (22).
2.5. BioEcon
BioEcon is a tool for epidemiologists facing decision about control measures. It automatically generates decision models of control strategies, which can then be compared interactively by a user. We are developing BioEconunder funding from the National Library of Medicine.
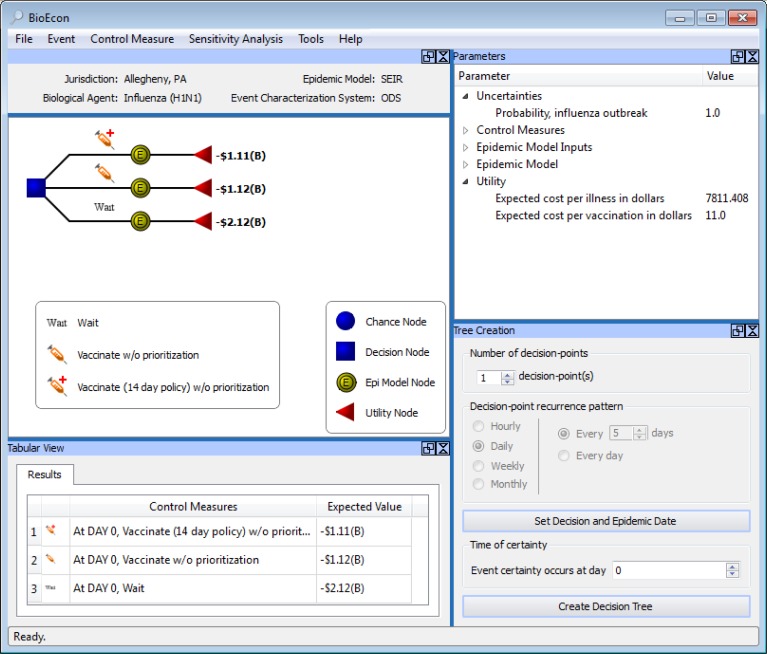
BioEcon generates a decision model from a set of control measures, an epidemic model, and a utility function (Figure 4). The square in the decision tree in Figure 4 represents a decision among three control measures. The three arcs from the decision node represent the three control measures. The deterministic nodes (double lined yellow circles with the letter ‘E’) represent three SEIR models for influenza, and the triangles represent the utility function.
Figure 4.
BioEcon.
In Figure 4, the three epidemic models use the same Ro, infectious period, and incubation period. However, the differential effects of the control measures can result in different initializations of the compartments and transition probabilities in the epidemic models. For example, the vaccination control measure adds direct transitions from the susceptible compartment to the recovered compartment.
BioEcon obtains the information needed to initialize the epidemic model either from ODS or an end user. When used with ODS, BioEcon computes the expected utility of each control measure (or sequence/combination of control measures) by model averaging over the set of SEIR models produced by ODS. Note that Figure 4 shows the expected utilities for a single ODS model.
An automatically generated decision model for influenza, Allegheny County, Sept 8, 2009 (retrospective analysis). The upper panel, left, shows the generated decision tree. Beneath it is a tabular display of the expected utilities (labelled ‘Expected Values’) of the decision alternatives. The utility function is: v(−$11) + s(−$7811.41), where v is number of people vaccinated, −$11 is the cost of vaccination, s is number sick, and −$7811.41 is the average cost per sick person, which equals the cost of illness and the loss of productivity.
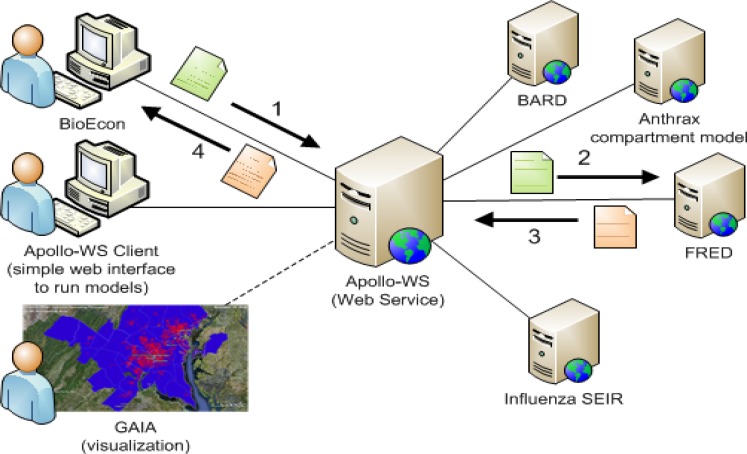
2.6. Apollo Web Service
We developed the Apollo Web Service to enable BioEconto connect to epidemic simulators developed by ourselves and others(Figure 5) and to also make out epidemic simulators available to other applications. At present, BioEcon can access four epidemic simulators through Apollo: a SEIR compartment model capable of modeling vaccination control measures; an influenza agent-based model capable of modeling vaccination and school closure interventions (among others); an aerosol release compartment model capable of modeling antibiotic prophylaxis control measures; and the BARD aerosol release simulator.
Figure 5.
Apollo Web Service
A simple end-user application that demonstrates the basic functionality of the Apollo Web Service can be found at http://research.rods.pitt.edu/apollo/.
End-user applications like BioEcon submit configuration objects to an epidemic simulator and receive output objects containing the results of the simulation (e.g., an epidemic curve).
3. Knowledge Bases
CDS, ODS, and BioEcon are knowledge-based systems. Aknowledge base is a component in a decision-support system that contains information that an expert might use to solve a problem in computer-interpretable format. It is standard practice to separate this knowledge from the other parts of a system, such as the inference algorithm that operates on the knowledge, to make this information more easy to maintain and verify(23).
The knowledge bases for CDS, ODS, and BioEconare depicted in Figure 1 as hexagons labeled “Disease models,” “Epidemic models,” and “Control measures and costs.”
Disease models, as previously discussed, use Bayesian networks to represent medical diagnostic knowledge—the symptoms, signs, and laboratory tests that a physician would use to diagnose a disease. The disease models represent the same kind of information that one finds in public health case definitions. A key difference is that these disease models are computer-readable and also include the sensitivity and specificity of each diagnostic finding for its disease. The disease models are also discussed in the accompanying paper in this issue of OJPHI.
Epidemic models represent expert knowledge about outbreaks. For example, a SEIR model represents influenza dynamics using a state transition network whose compartments represent disease states (e.g., susceptible and infectious) and whose parameters (e.g., infectious period, latent period)specify the rates at which transitions between disease states states over time.
Control measures and costs: BioEcon represents knowledge that expert epidemiologist use when making decisions about control measures. BioEcon represents this knowledge using objects, which are nested structures that can inherit characteristics from more general parent classes. The object representing a vaccination control measure, for example, has the following attributes: jurisdiction (e.g., Allegheny County), supply schedule, vaccine administration capacity, efficacy, and lists of other control measures that it can run concurrent with, follow, or precede. BioEcon can acquire and store this information for multiple jurisdictions, each of which can have different capacities.
BioEcon also requires cost information, including cost of illness, lost productivity, which is knowledge on which rationale decision-making is based. We have not developed extensive representations of cost information within BioEcon to date; instead, we use Excel spreadsheets and other tools to develop detailed economic models, and represent the rolled up costs in BioEcon as components of its utility functions.
4. An Example of Information Flow and Transformation
This section shows the flow of information through the components of the probabilistic, decision-theoretic disease surveillance and control system using influenza as an example. In particular, it shows the information that is passed in the rounded boxes in Figure 1 labeled Coded visit data, Differential diagnosis, Population analyses, and Decision analyses. We assume the reader is familiar with the patient data stored in electronic form in EMRs; therefore, we begin with the input to CDS, which are patient findings in standard format.
Coded visit data. The following table shows the coded patient findings obtained by CDS from Phoenix for three ED visits on September 1, 2009. Approximately 600 patients were seen that day in monitored EDs.
| Patient visit | UMLS Concept ID | Name of finding | Value |
|---|---|---|---|
| A | C0000729 | Abdominal pain | Absent |
| C0015967 | Fever | Absent | |
| C0018681 | Headache | Present | |
| C0027497 | Nausea | Present | |
| C0043144 | Wheezing | Absent | |
| C0085593 | Chills | Absent | |
| C1883552 | Weakness | Absent | |
| B | C0015967 | Fever | Absent |
| C0085593 | Chills | Absent | |
| C1883552 | Weakness | Absent | |
| C | C0015672 | Fatigue | Absent |
| C0015967 | Fever | Absent | |
| C0085593 | Chills | Absent | |
| C1883552 | Weakness | Absent |
Differential diagnosis of patients. The following table shows the output sent by CDS to ODS for three patient visits on September 1, 2009 in monitored EDs. They are not the same patients visits as in the previous table.
| Patient | P (evidence of patient | influenza) | P (evidence of patient | not influenza) |
|---|---|---|
| 1 | 0.015759230220164264 | 0.0040469980953649898 |
| 2 | 1.3756216427857736e-008 | 1.754268236767679e-005 |
| 3 | 0.00047169035934720448 | 0.020177483460983615 |
Population analyses. The following information represents the output sent by ODS to BioEcon for Sept 1, 2009. We are showing numerical output, however, the results are amenable to graphical display as well. We can plot the posterior probability of an influenza outbreak, epidemic curves, and other quantities of interest.
start monitoring date: May 28, 2009
current date: September 1, 2009
prior probability that an ED patient has influenza on the current day: 0.007
number of [SEIR] models searched: 50,000
total run time: 737.3 seconds
// Outbreak Detection //
posterior probability of an influenza outbreak: 0.549 [The number relevant to outbreak detection]
//Outbreak Characterization//
- [the following output is for one of 50,000 SEIR models searched—the full output from ODS contains all 50,000]
- model_posterior_probability: 0.000113
- S: 1215434 (number of individuals in susceptible compartment on September 1, 2009)
- E: 601 (exposed)
- I: 903 (infectious)
- R: 1652 (recovered)
- latent_period: 2.6 days
- infectious_period: 5.9 days
- Ro: 1.855
- Outbreak start day: 64 days after the start of monitoring on May 28, 2009
- Initial number infected: 96
Decision analysis. Figure 4 shows the output of a decision analysis, which is a base case analysis with sensitivity analyses (not shown). For expository purposes, the following table shows the mathematical result of a decision model, which is the set of expected utilities computed for the decision alternatives analyzed by the model. The control measure recommended by the decision model in this example is vaccination, with aggressive and routine administration differing by less than 1%.
| Decision alternative (control measure) | Expected utility |
|---|---|
| Vaccinate 14-day policy, without prioritization | −$1.11B |
| Vaccinate without prioritization (routine vaccination) | −$1.12B |
| No vaccination | −$2.12B |
5. Significance
In 1959, Ledley and Lusted identified probability and decision theory as the mathematical foundations of medical diagnosis (24). This insight has had significant impact on the fields of clinical medicine, medical decision-making, and computerized medical decision support.
In 2001, we observed that probability and decision theory were also the mathematical foundations of disease surveillance and control (25).Further, the methods developed for probabilistic medical diagnosis could be applied, without modification, to case detection for disease surveillance. We observed that probability and decision-theory were an ideal basis for representation and inference at the population level, and that the domains of medical diagnosis and population diagnosis could be bridged by these formalisms.
But despite the potential of a probabilistic decision-theoretic approach, the practice of disease surveillance still rests on a Boolean foundation: For analytic purposes, a population is represented by 1’s and 0’s, where 1 denotes that an individual has a disease of interest, and 0 indicates that the disease status of the individual is unknown. This limitation applies not only to conventional disease reporting, but also to electronic laboratory reporting and syndromic surveillance.
A fundamental problem with a Boolean foundation for disease surveillance is that a yes–no classification of a patient into disease or no disease is an information-losing first step in a process that requires maximum use of information as it becomes available (26). It cannot represent the level of uncertainty about a patient’s diagnosis except by ad hoc extensions such as the diagnoses “suspected SARS” and “probable SARS.” It cannot integrate data from syndromic surveillance, ELR, and other disease surveillance paradigms to form a more confident assessment of a patients disease state. The net effect is case and outbreak detection and characterization are less sensitive, specific, and timely than they could be.
Effectively integrating Bayesian medical diagnosis and population diagnosis will address critical barriers to progress in both fields, and can open up major avenues for new research and real-world applications in clinical medicine and public health.
In clinical medicine, for example, the answer to the often-voiced criticism, “Where do you get the priors?” will be: “From real-time population level analysis.” This problem is particularly nagging for outbreak diseases, where the prior probability may change quickly. A solution to this problem will make case detection more timely, sensitive, and specific for outbreak diseases, with practical benefits for hospital infection control, quality assurance, and case detection for disease surveillance.
In public health, the biggest advantage of the probabilistic, decision-theoretic approach is that it is a well-organized and formally sound method for integrating multiple weak signals, with medical knowledge, and with epidemiological knowledge, to provide for early and reliable detection at low false alarm rates. Also significant, is our approach to the problem of how to synchronize epidemic models with an actual population, which will make epidemic models more useful during outbreaks.
Integrating probabilistic disease surveillance and medical diagnosis is a paradigm shift for public health. Bayesian diagnosis of individual patients is not yet a component in future envisioned architectures such as the Public Health Information Network(27), nor is it factored into CMMS “meaningful use,” which is incentivizing the healthcare system to modify their information systems in other ways. The architectures, message types, and data standards currently under development will not be able to support this paradigm, unless they are designed with its requirements in mind.
The limitations of the current Boolean paradigm will become increasingly problematic as disease surveillance takes fuller advantage of data stored in EMRs.
6. Future Work
At present, the probabilistic, decision-theoretic system monitors influenza in Allegheny County. The CDS component is located in a machine room in the UPMC health system. ODS, BioEcon, and three epidemic models are running on servers in our laboratory as Web Services. We consider the current implementation an operational prototype.
Our future work will measure the performance of CDS and ODS for influenza and test hypotheses about the performance synergies achieved as a result of their integration. We also plan to expand the number of diseases being modeled.
Acknowledgments
The Centers for Disease Control and Prevention 1P01HK000086-01 supports the “University of Pittsburgh Center for Advanced Study of Informatics.” The National Library of Medicine grant NLM 5R01LM009132-02 “Decision Making in Biosurveillance” supports development of BioEcon and the Apollo Web Service. The authors would like to thank Lee Husting, MD for his encouragement and professional dedication.
References
- 1.Zimmer SM, Crevar CJ, Carter DM, Stark JH, Giles BM, Zimmerman RK, et al. Seroprevalence following the second wave of Pandemic 2009 H1N1 influenza in Pittsburgh, PA, USA. PLoS One. 2010;5(7):e11601–e. doi: 10.1371/journal.pone.0011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burkom HS, Ramac-Thomas L, Babin S, Holtry R, Mnatsakanyan Z, Yund C. An integrated approach for fusion of environmental and human health data for disease surveillance. Statistics in Medicine. 2011;30(5):470–9. doi: 10.1002/sim.3976. [DOI] [PubMed] [Google Scholar]
- 3.Harvey AC. The Kalman filter and its applications in econometrics and time series analysis. Methods of Operations Research. 1982;44(1):3–18. [Google Scholar]
- 4.Le Strat Y, Carrat F. Monitoring epidemiologic surveillance data using hidden Markov models. Statistics in medicine. 1999;18(24):3463–78. doi: 10.1002/(sici)1097-0258(19991230)18:24<3463::aid-sim409>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 5.Mnatsakanyan ZR, Burkom HS, Coberly JS, Lombardo JS. Bayesian information fusion networks for biosurveillance applications. Journal of the American Medical Informatics Association. 2009;16(6):855–63. doi: 10.1197/jamia.M2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nobre FF, Monteiro ABS, Telles PR, Williamson GD. Dynamic linear model and SARIMA: a comparison of their forecasting performance in epidemiology. Statistics in medicine. 2001;20(20):3051–69. doi: 10.1002/sim.963. [DOI] [PubMed] [Google Scholar]
- 7.Ong JBS, Chen MIC, Cook AR, Lee HC, Lee VJ, Lin RTP, et al. Real-Time Epidemic Monitoring and Forecasting of H1N1-2009 Using Influenza-Like Illness from General Practice and Family Doctor Clinics in Singapore. PLoS ONE. 2010;5(4):e10036. doi: 10.1371/journal.pone.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proceedings of the IEEE. 1989;77(2):257–86. [Google Scholar]
- 9.Rath T, Carreras M, Sebastiani P. Automated detection of influenza epidemics with hidden Markov models. Advances in Intelligent data analysis V. 2003:521–32. [Google Scholar]
- 10.Sebastiani P, Mandl KD, Szolovits P, Kohane IS, Ramoni MF. A Bayesian dynamic model for influenza surveillance. Statistics in Medicine. 2006;25(11):1803–16. doi: 10.1002/sim.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiryaev AN. Optimal stopping rules. Springer; 1978. [Google Scholar]
- 12.Stroup DF, Thacker SB. A Bayesian approach to the detection of aberrations in public health surveillance data. Epidemiology. 1993:435–43. doi: 10.1097/00001648-199309000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Watkins R, Eagleson S, Veenendaal B, Wright G, Plant A. Disease surveillance using a hidden Markov model. BMC medical informatics and decision making. 2009;9(1):39. doi: 10.1186/1472-6947-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neill DB, Cooper GF. A multivariate Bayesian scan statistic for early event detection and characterization. Machine Learning. 2010;79(3):261–82. [Google Scholar]
- 15.Wong W-K, Moore A, Cooper G, Wagner M, editors. Bayesian Network Anomaly Pattern Detection for Disease Outbreaks; Proceedings of the Twentieth International Conference on Machine Learning; Menlo Park. California: AAAI Press; 2003. [Google Scholar]
- 16.Cooper GF, Dash DH, Levander JD, Wong W-K, Hogan WR, Wagner MM, editors. Bayesian biosurveillance of disease outbreaks; Proceedings of the Twentieth Annual Conference on Uncertainty in Artificial Intelligence; Banff, Canada: AUAI Press; 2004. 1036855: [Google Scholar]
- 17.Jiang X, Cooper GF. A Bayesian spatio-temporal method for disease outbreak detection. Journal of the American Medical Informatics Association. 2010;17(4):462. doi: 10.1136/jamia.2009.000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan T-C, King C-C, Yen M-Y, Chiang P-H, Huang C-S, Hsiao CK. Probabilistic Daily ILI Syndromic Surveillance with a Spatio-Temporal Bayesian Hierarchical Model. PLoS ONE. 2010;5(7):e11626. doi: 10.1371/journal.pone.0011626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong JB, Chen MI, Cook AR, Lee HC, Lee VJ, Lin RT, et al. Real-time epidemic monitoring and forecasting of H1N1-2009 using influenza-like illness from general practice and family doctor clinics in Singapore. PLoS One. 2010;5(4):e10036. doi: 10.1371/journal.pone.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skvortsov A, Ristic B, Woodruff C, editors. Predicting an epidemic based on syndromic surveillance; Proceedings of the Conference on Information Fusion; 2010. [Google Scholar]
- 21.Vynnycky E, White R. An introduction to infectious disease modelling. Oxford University Press; 2010. [Google Scholar]
- 22.Hogan WR, Cooper GF, Wallstrom GL, Wagner MM, Depinay JM. The Bayesian aerosol release detector: An algorithm for detecting and characterizing outbreaks caused by an atmospheric release of Bacillus anthracis. Statistics in Medicine. 2007;26(29):5225–52. doi: 10.1002/sim.3093. [DOI] [PubMed] [Google Scholar]
- 23.Buchanan BG, Smith RG. Fundamentals of Expert Systems. Annu Rev Comput Sci. 1988;3:23–58. [Google Scholar]
- 24.Ledley RS, Lusted LB. Reasoning foundations of medical diagnosis. Science. 1959;130(3366):9–21. doi: 10.1126/science.130.3366.9. [DOI] [PubMed] [Google Scholar]
- 25.Wagner M, Tsui F, Espinio J, Dato V, Sittig D, Caruana R, et al. The emerging science of very early detection of disease outbreaks. J Public Health Manag Pract. 2001;7(6):51–9. doi: 10.1097/00124784-200107060-00006. [DOI] [PubMed] [Google Scholar]
- 26.Wagner M, Wallstrom GL. Chapter 1 Introduction. In: Wagner M, Moore A, Aryel R, editors. Handbook of Biosurveillance. New York: Elsevier; 2006. [Google Scholar]
- 27.CDC . Public Health Information Network (PHIN) Strategic Plan (draft version 2.2.1) Atlanta: CDC; Mar 17, 2011. [Google Scholar]
- 28.Neill DB, Moore AW, Cooper GF. A Bayesian Spatial Scan Statistic. Advances in Neural Information Processing Systems. 2006;18:1003–10. [Google Scholar]