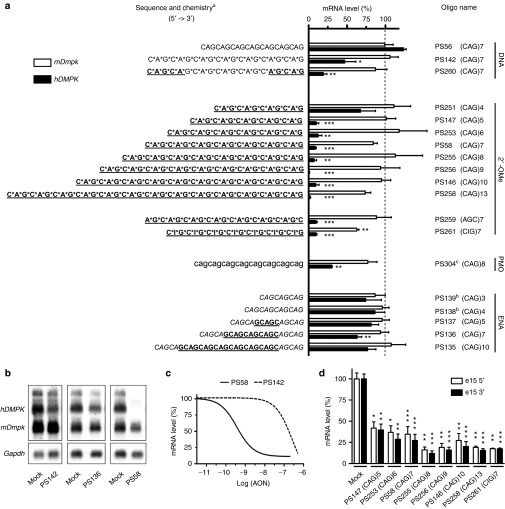
Figure 1.
Collection of triplet repeat AONs tested for their ability to silence expanded hDMPK mRNA in DM500 myotubes. (a) Summary of silencing efficiency. Endogenous mDmpk RNA was included as a negative control, since it lacks a pure (CUG)n repeat. AONs are grouped according to their main chemical modification: aDNA (capital); RNA (capital, underlined); PT, phosphorothioate linkage (*); 2′-OMe, 2′-O-methyl sugar modification (capital, underlined, bold); ENA, 2′-O,4′-C-ethylene-bridged nucleic acid (capital, italics); PMO, morpholino phosphorodiamidate (lower case); bCy3-labeled; cCoupled to octaguanidine dendrimer. The means of at least three independent experiments for each AON are shown. Dashed line indicates 100% levels (mock samples). (b) Representative northern blots with RNA isolated from AON-treated DM500 myotubes probed with a hDMPK and a Gapdh probe. Results of treatment with three AONs (200 nmol/l) or mock treatment is shown. (c) Concentration–response curves of PS58 and PS142. DM500 myotubes were treated with a concentration series of 0.01–500 nmol/l AON (see also Supplementary Figure S2). (d) RT-qPCR analysis of hDMPK RNA levels in DM500 myotubes after treatment with a selection of 2′-OMe AONs (200 nmol/l, n ≥ 3). PCR amplicons were located in exon 15, either 5′ or 3′ to the (CUG)n tract. *P < 0.05; **P < 0.01; ***P < 0.001. AON, antisense oligonucleotide; Gapdh, glyceraldehyde 3-phosphate dehydrogenase; hDMPK, human DM protein kinase; mDmpk, mouse Dmpk; RT-qPCR, reverse transcription-quantitative PCR.