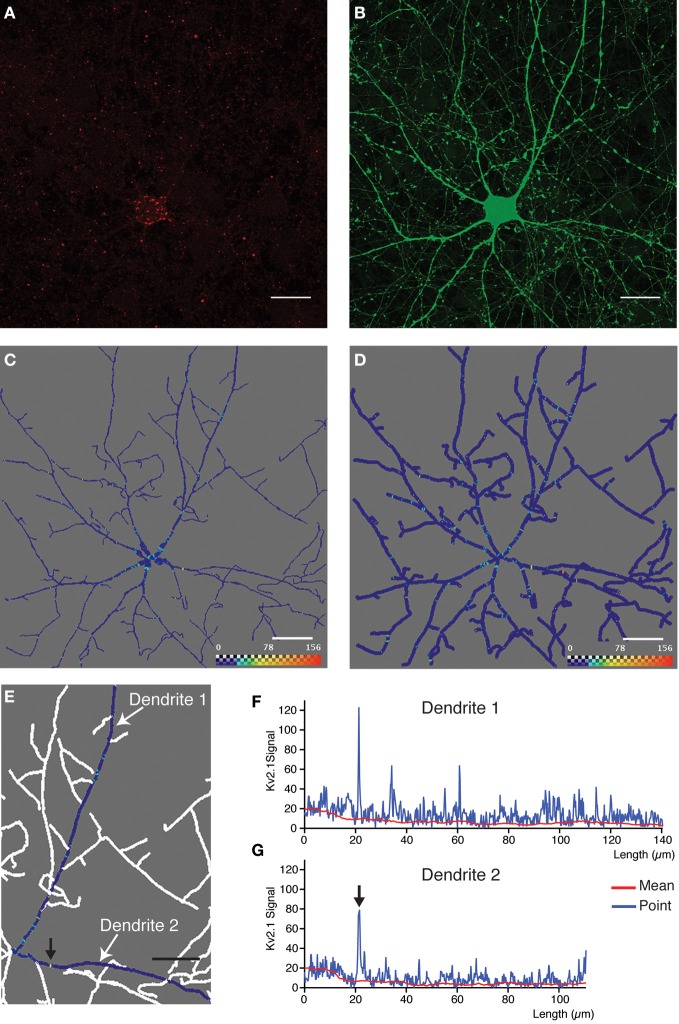
Figure 4.
Quantitative analysis and 3D profiling of endogenous Kv2.1 signal in cultured neocortical neurons. (A,B) Image stacks corresponding to endogenous Kv2.1 signal (A) and morphological label (eGFP expression) (B). Images are z-projections derived from maximum signal intensity across 36 focal planes generated from data that has been subjected to 2 rounds of blind deconvolution. (C) Two-dimensional snapshot taken from Amira viewer window showing neuronal reconstruction overlaid with 3D ion channel profile derived from point fluorescence values calculated along the dendrite skeleton. The color map has been adjusted to best represent, on a graphical level, the predominant ion channel clusters. Scale bar: 20 μm. In this case, the skeleton is represented as a true morphological skeleton (incorporating information about the radius of the dendrite). (D) Two-dimensional snapshot taken from Amira viewer window (same dataset as in C) showing reconstructed skeleton (represented as a filament). (E) Magnification of a section of the reconstructed skeleton, showing 3D ion channel profile associated with two individual dendrites (arbitrarily named dendrite 1 and dendrite 2). Here, point fluorescence's values are presented by means of the color map function. The color map has been adjusted to best represent, on a graphical level, the predominant ion channel clusters. (F,G) Fluorescence intensity plotted as a function of dendritic length for the same dendrites. Data is presented as the point fluorescent value as well as mean fluorescent value calculated at each point. Arrows in (E) and (G) indicate a putative ion channel cluster, clearly visible in both representations of the data. Scale bar, 20 μm.