Figure 1.
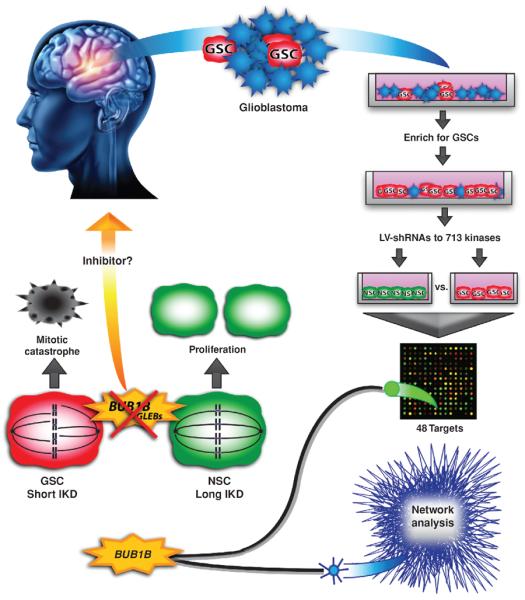
Schematic of experimental flow and results as described by Ding and colleagues (6). Primary human glioblastoma cells were taken from resected tumors and grown in conditions that enrich for GSCs. These cells, along with NSCs, were infected with a shRNA library that targeted the human kinome. Both cell types were then expanded for 21 days before isolating RNA and submitting it for microarray. Microarray analysis revealed 48 candidate targets that were depleted in GSCs, but not fetal NSCs. Network analysis was done on The Cancer Genome Atlas (TCGA) expression data to inform the results of the shRNA screen. By combining both methods, the authors were able to identify BUB1B/BubR1 as their top candidate. BUB1B/BubR1 is a protein that monitors proper spindle microtubule attachment to the kinetochore, and whose GLEBs domain is specifically required when the IKD is short, as is the case in GSCs. Knockdown of BUB1B/BubR1 in cells with a short IKD leads to mitotic catastrophe and cell death. However, in NSCs the IKD is longer and BUB1B/BubR1 is not required in this process, allowing for normal proliferation even in the presence of BUB1B/BubR1 knockdown. This feature makes BUB1B/BubR1, or potentially other proteins involved in this process, promising candidates for targeted therapies.
