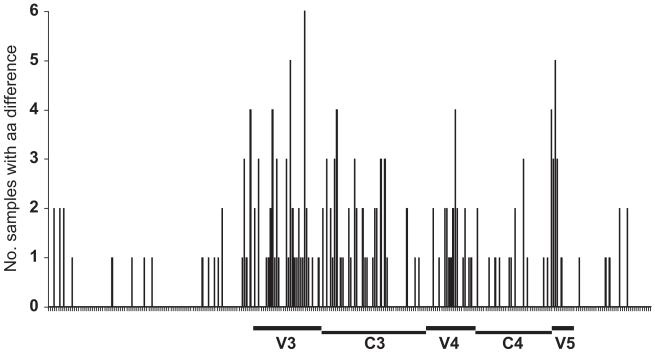
Fig. 5.
All env sequences were aligned, and V1–V2, V3, V4–V5 segments corresponding to HXB2 env amino acids were defined. Signature amino acid differences between intra-patient R5 and CXCR4-using clones were mapped relative to the HXB2 env numbering. Signature amino acids were determined using the VESPA program with threshold of 0 and comparison of R5 and CXCR4-using sequences from within each of the 8 subjects. Two DM samples which were not composed of R5 clones, with only X4 and dual-tropic clones, were not included in the analysis.