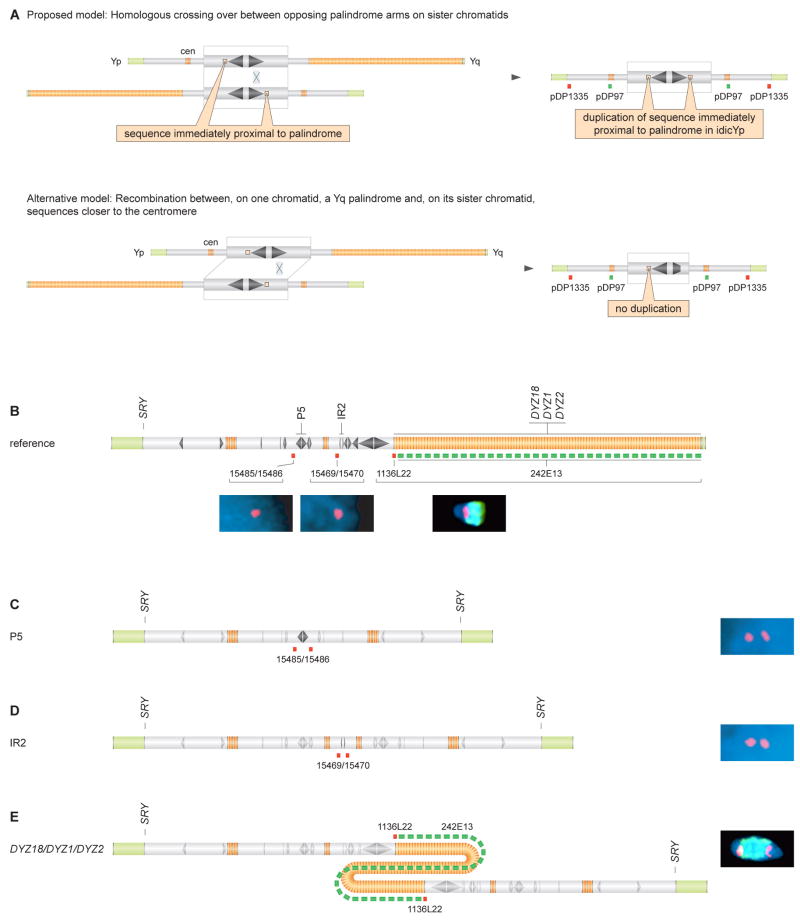
Figure 4. Duplication in idicYp chromosomes of sequences immediately proximal to targeted palindromes.
(A) Two models of idicYp formation can be distinguished by determining copy numbers of sequences immediately proximal to targeted palindrome.
(B) Probe hybridization sites shown below chrY schematic. Hybridizations to chrY of normal male produced expected signals; 15485/15486 and 15469/15470 on interphase spreads, and co-hybridization of 1136L22 and 242E13 on metaphase spreads.
(C) Interphase FISH demonstrates duplication of 15485/15486 on idicYp with breakpoint in palindrome P5.
(D) Interphase FISH demonstrates duplication of 15469/15470 on idicYp with breakpoint in inverted repeat IR2.
(E) Metaphase FISH demonstrates duplication of 1136L22 on idicYp with breakpoint in DYZ18/DYZ1/DYZ2 heterochromatin. Co-hybridization of 1136L22, in red, with 242E13, in green, produced predicted red-green-red pattern.
All micrographs at same magnification. In each interphase FISH experiment, ≥100 nuclei were scored (Figures S3, S4, and S5).