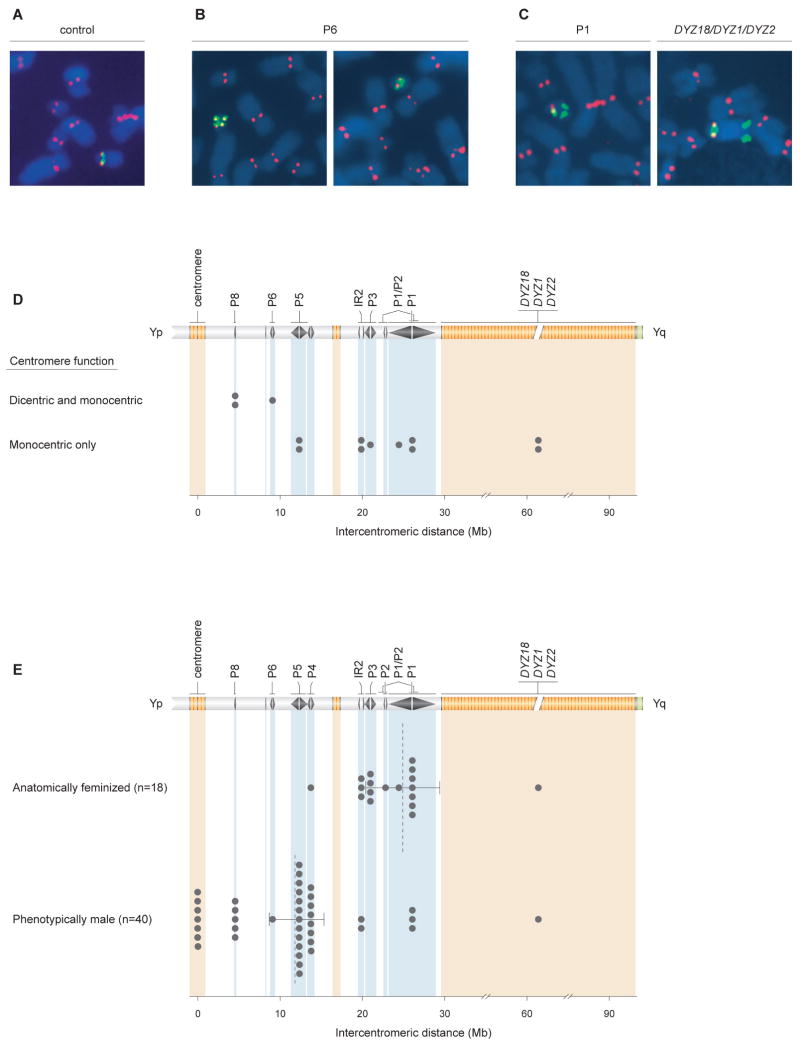
Figure 6. Correlates of intercentromeric distance in idicYp chromosomes: centromere inactivation and sex reversal.
(A) Active chrY centromeres identified by immunofluorescence followed by FISH (IF-FISH). Anti-CENP-E antibody staining, in red, identifies all active centromeres. FISH probe pDP97 (to DYZ3 alpha satellite repeats), in green, identifies chrY centromeric DNA. IF-FISH to chrY of normal male control produced expected signals at single centromere.
(B) IF-FISH to idicYp case with breakpoint in palindrome P6 demonstrates cell-to-cell mosaicism for functionally dicentric (left) or functionally monocentric (right) chromosomes.
(C) IF-FISH to idicYp cases with breakpoints in palindrome P1 (left) or in DYZ18/DYZ1/DYZ2 heterochromatin (right) demonstrates functional inactivation of one Y centromere.
(D) Plots of recombination sites in 13 idicYp cases assayed by IF-FISH and displaying either cell-to-cell mosaicism for functionally dicentric and monocentric idicYp chromosomes (three cases, above) or exclusively functionally monocentric idicYp chromosomes (ten cases, below). For each cell line, ≥20 idicYp-bearing spreads of good morphology were scored. Among the three cases with cell-to-cell variation, fraction of cells displaying functionally dicentric idicYp chromosomes ranged from 41% to 77%. See Figure S8 for additional data.
(E) Plots of intercentromeric distances in SRY-positive idicYp or isoYp chromosomes in 18 feminized individuals (above) and 40 males (below). Dotted lines indicate mean intercentromeric distances for the two groups; 95% confidence intervals calculated by bootstrapping method.