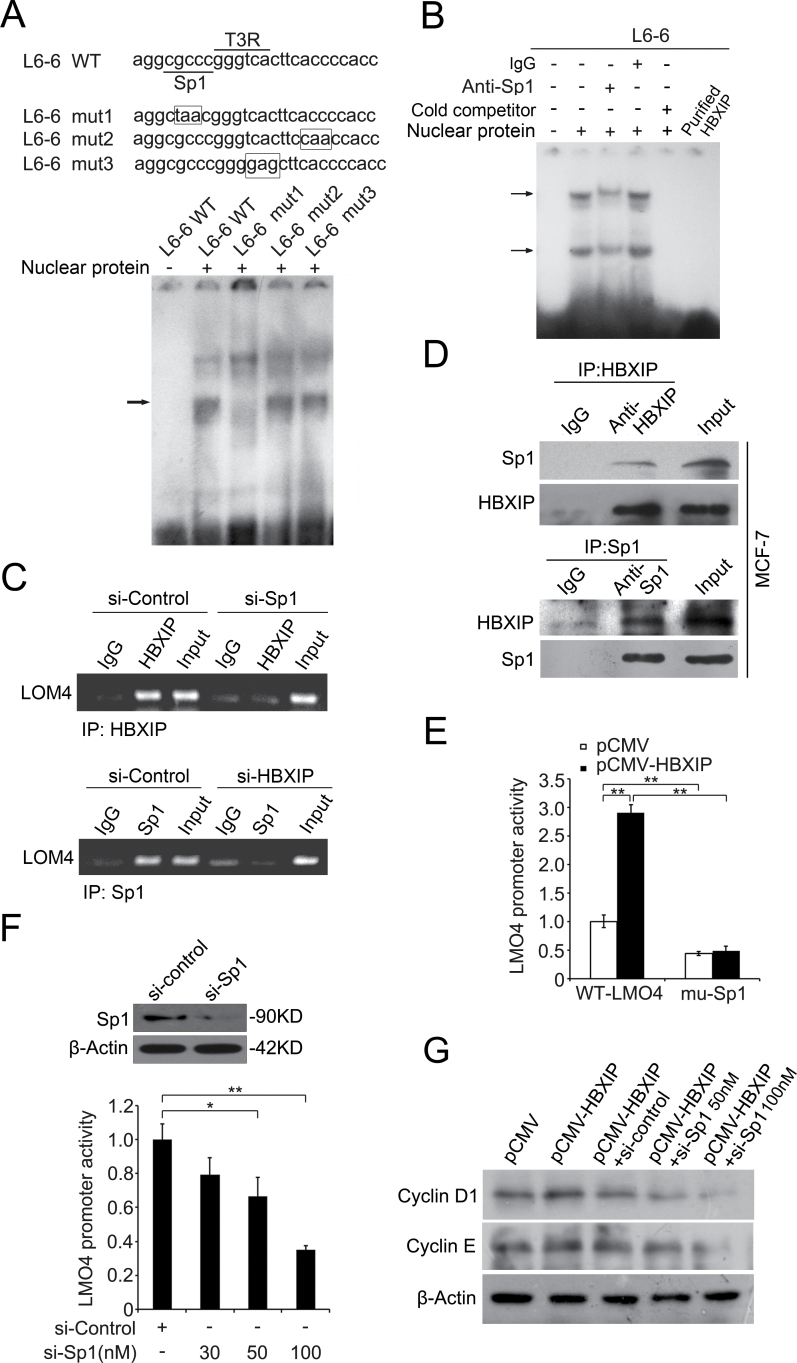
Fig. 4.
HBXIP activates LMO4 promoter through transcription factor Sp1. (A) A model of wild and mutation type analysis for the fragments of L6-6 (-237/-206). The binding sites of Sp1and T3R were underlined. The mutated nucleotides of both transcription factor recognizing sequences as well as the mutation of 3'-flank region of L6-6 fragment were highlighted with rectangles. EMSA showed the interaction of the above DNA probes with nucleus proteins of MCF-7 cells. The L6-6 mut 1 dismissed the interaction. (B) EMSA with the addition of anti-Sp1 antibodies was performed to examine the binding of nucleus proteins to fragment L6-6. (C) ChIP showed the interaction of HBXIP with LMO4 promoter in MCF-7 cells when Sp1 was knockdown, and vice versa. (D) The interaction between HBXIP and Sp1 was determined by co-immunoprecipitation in MCF-7 cells. The immunoprecipitates were analyzed by western blot. (E) The promoter activity of LMO4 mediated by HBXIP was measured by luciferase reporter gene assay when Sp1 binding site was mutated. MCF-7-pCMV and MCF-7-HBXIP cells were transiently transfected with pGL3-WT-LMO4 or pGL3-mu-Sp1 plasmids (**P < 0.01; Student’s t-test). (F) The promoter activity of LMO4 was examined by luciferase reporter gene assay in MCF-7-HBXIP cells when the cells were treated with Sp1 siRNA (30, 50 or 100nM) (*P < 0.05, **P < 0.01; Student’s t-test), in which the siRNA efficiency of Sp1 was determined by western blot analysis. (G) The expression levels of cyclin D1 and cyclin E were examined by western blot in MCF-7 cells transiently transfected with pCMV-HBXIP (or pCMV-tag2B), pCMV-HBXIP and Sp1 siRNA (or control siRNA), respectively.