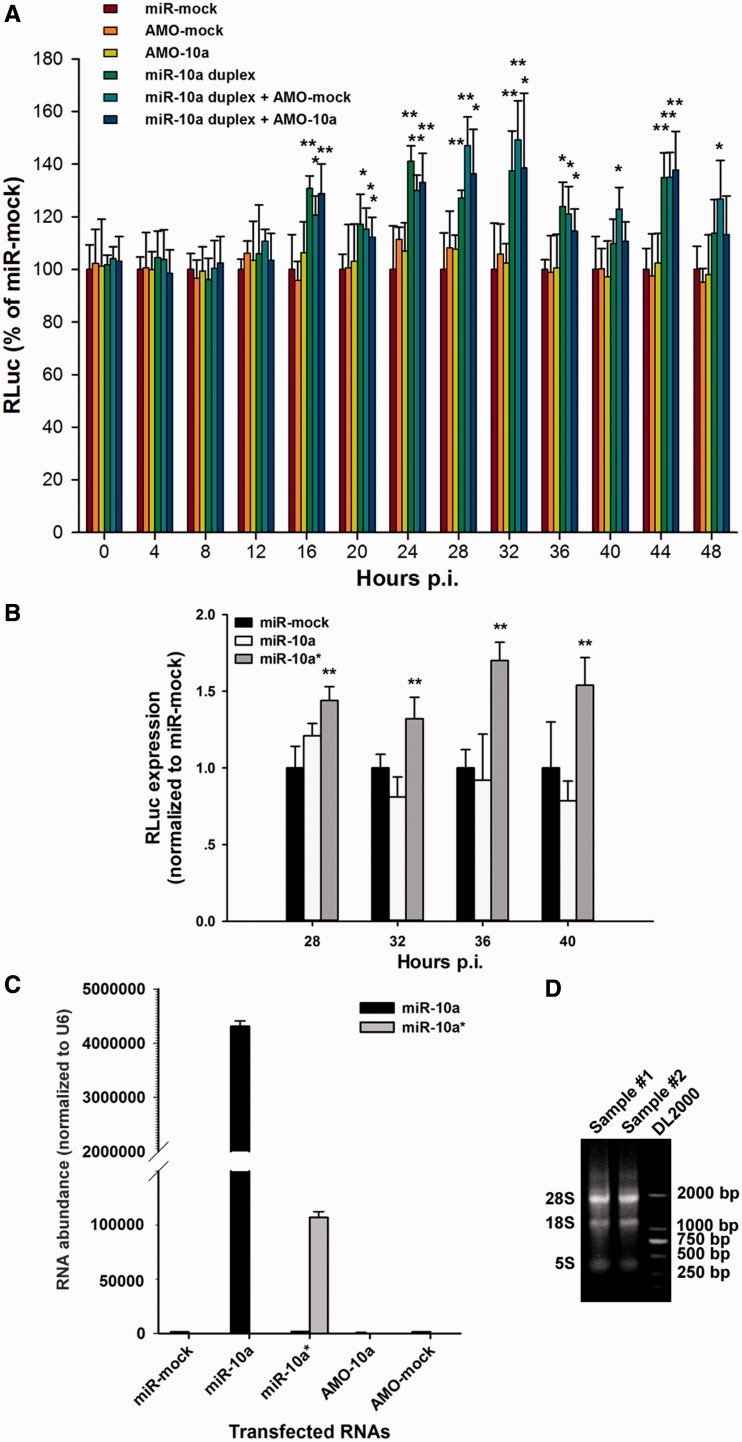
Figure 2.
MiR-10a* up-regulates the biosynthesis of CVB3 in HeLa cells. (A) HeLa cells were transfected with miR-mock, AMO-mock, AMO-10a, miR-10a duplex, miR-10a duplex + AMO-mock and miR-10a duplex + AMO-10a. The cells were then infected with RLuc-CVB3 (MOI = 0.01), and harvested for RLuc detection at 4-h intervals from 0 to 40 h p.i. The RLuc level was normalized to that of the miR-mock-treated cells. Error bars represent SDs (n = 6). (B) HeLa cells were transfected with single-stranded miR-10a* or single-stranded miR-10a as described above. The cells were then infected with RLuc-CVB3 (MOI = 0.01), and harvested for RLuc detection at 4-h intervals from 28 to 40 h p.i. The RLuc level was normalized to that of the miR-mock-treated cells. Error bars represent SDs (n = 6). (C) The transfected RNA abundance in the treated HeLa cells. HeLa cells were transfected with miR-mock, single-stranded miR-10a, single-stranded miR-10a*, AMO-10a and AMO-mock. The miR-10a and miR-10a* abundance was detected by RT-qPCR at 24 h posttransfection and normalized to the U6 snRNA abundance in the cells. Error bars represent the SDs (n = 3). (D) The integrity of RNA extraction of HeLa cells. The total RNA was extracted from the transfected HeLa cells with TRIzol reagents. About 2.5 μg of RNA (sample #1 and #2) and 1 μg of DNA ladder DL2000 were loaded to 0.7% agarose gel for electrophoresis. The ratio of 28S/18S RNA was ∼1.8. * and ** represent P < 0.05 and P < 0.01, respectively, compared with the miR-mock-treated group by Student’s t-test.