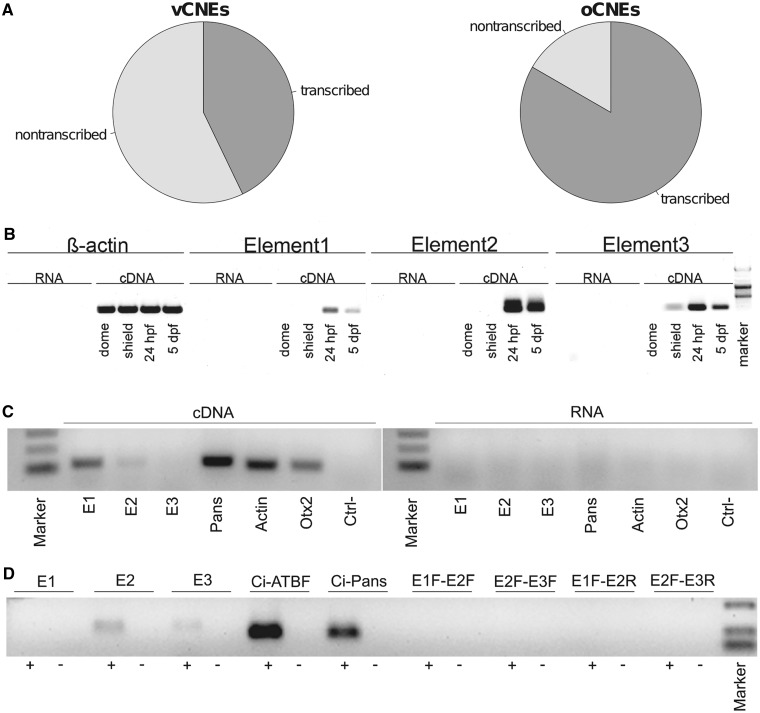
Figure 3.
Potential transcription of oCNEs: The oCNEs result to be enriched for eRNAs. Pie-charts in (A) show how vCNEs and oCNEs overlapping enhancers from Kim et al. (49) segregate between the classes of eRNAs and non-transcribed enhancers. Twenty-eight vCNEs overlap enhancers from Kim et al. and ∼40% of them overlap eRNAs. Conversely, 18 oCNEs overlap enhancers from Kim et al. and >80% of them overlap eRNAs. The three oCNEs used for the validation of the enhancer function were also validated for transcriptional activity in D. rerio (B), M. musculus (C) and C. intestinalis (D). Primers were designed to amplify a fragment of ∼100 bp around each element. As positive control, we used the following coding transcripts: bActin (D. rerio and M. musculus), Otx2 (M. musculus), Ci-atbf (C. intestinalis). Non-coding transcripts used were as follows: Ci-Pans (C. intestinalis) (51) and Pans (Mm.221244, the murine homolog of Ci-Pans) (52). All the used controls are known to be expressed at the time of the sampling. As negative control, we used DNAseI-digested RNA (indicated as RNA in B and C and as ‘-’ in D). In C. intestinalis, we also used different combinations of the forward/reverse primers. The absence of signal in the cDNA template PCR is indicative of the absence of genomic DNA contamination in the cDNA preparation demonstrating that the amplicons are real RNA products.