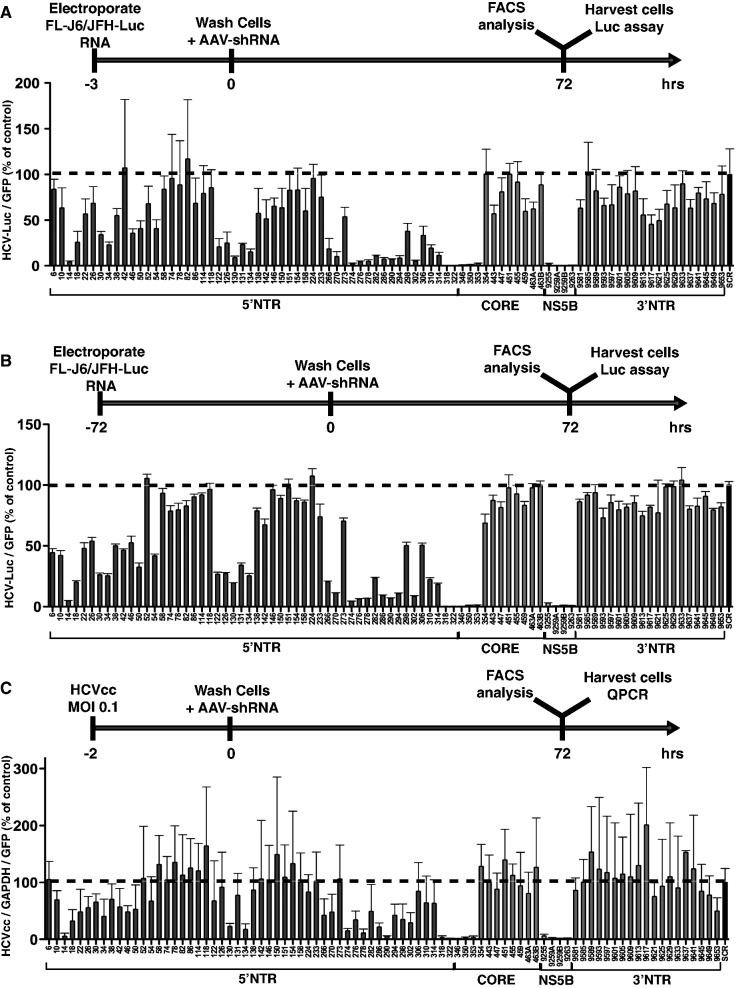
Figure 2.
All shRNAs were tested against replicating FL-J6/JFH-Luc HCV in a transient-replication model (A), a stable-replication model (B), as well as against HCVcc infection (C) in Huh7.5 cells. Upper portions of panels A–C show graphic representation of each experimental design. The number corresponding to each shRNA represents the position of the first nucleotide in the recombinant HCV J6(5′UTR-NS2)/JFH1 (GenBank: JF343782.1). The shRNA-SCR was used as negative control and was assigned a value of 100. In (A) and (B), FL-J6/JFH-Luc luciferase levels were normalized to AAV-shRNA encoded eGFP to account for transduction efficiency, whereas in (C), GAPDH quantitative PCR levels and AAV-shRNA eGFP levels were used to normalize the HCV RNA levels measured by quantitative PCR. Error bars represent SD from n = 3–6.