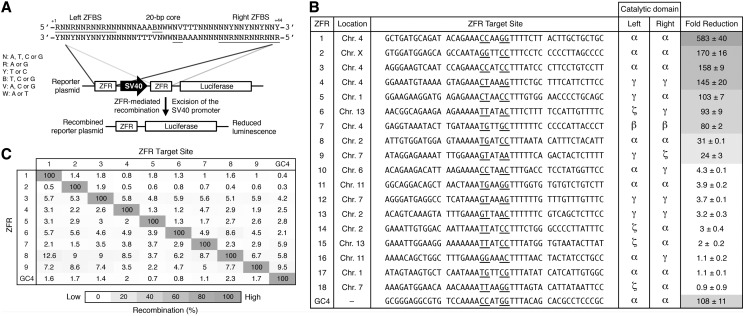
Figure 4.
ZFRs recombine user-defined sequences in mammalian cells. (A) Schematic representation of the luciferase reporter system used to evaluate ZFR activity in mammalian cells. ZFR target sites flank an SV40 promoter that drives luciferase expression. Solid lines denote the 44-bp consensus target sequence used to identify potential ZFR target sites. The consensus ZFR target site consists of two-inverted 12-bp ZFBS flanking a central 20-bp core sequence recognized by the ZFR catalytic domain. Underlined bases indicate zinc-finger targets and positions 3 and 2. (B) Fold-reduction of luciferase expression in HEK293T cells co-transfected with designed ZFR pairs and their cognate reporter plasmid. Fold-reduction was normalized to transfection with empty vector and reporter plasmid. Renilla luciferase expression was used to normalize for transfection efficiency and cell number. The sequence identity and chromosomal location of each ZFR target site and the catalytic domain composition of each ZFR pair are shown. Underlined bases indicate positions 3 and 2. Standard errors were calculated from three independent experiments. ZFR amino acid sequences are provided in Supplementary Table S3. (C) Specificity of ZFR pairs. Fold-reduction of luciferase expression was measured for ZFR pairs 1 through 9 and GinC4 for each non-cognate reporter plasmid. Recombination was normalized to the fold-reduction of each ZFR pair with its cognate reporter plasmid. Assays were performed in triplicate.