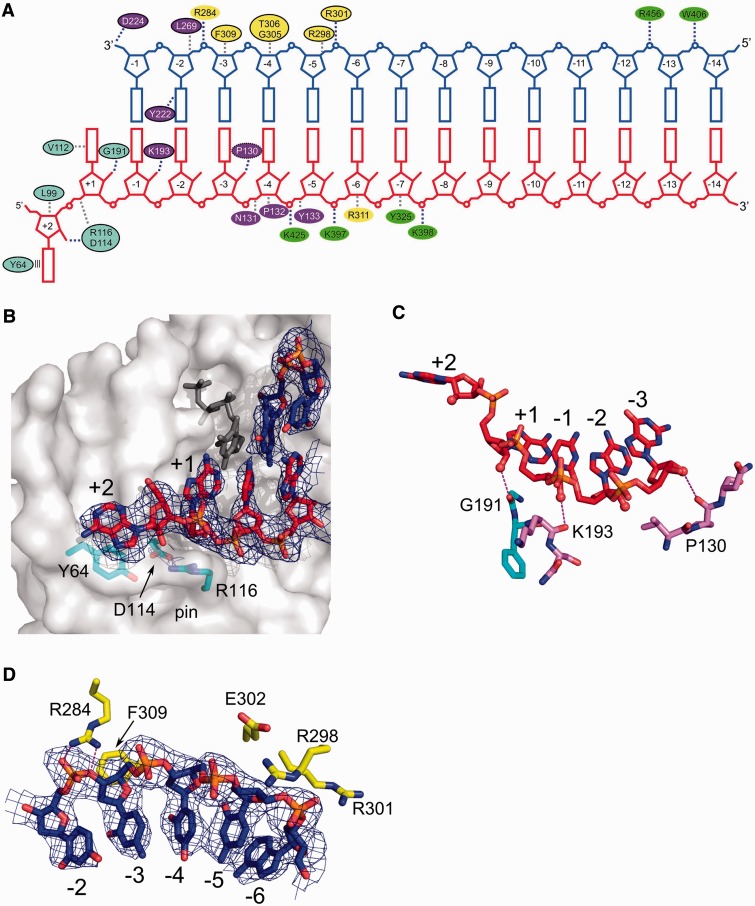
Figure 2.
Substrate binding. (A) Diagram showing interactions between XMRV RT and nucleic acid. Ovals are coloured according to protein subdomains of Figure 1. Black outline denotes residues for which equivalents can be found in HIV-1 RT (putative equivalent denoted with dashed oval). Stacking of Tyr64 with the RNA overhang is shown as parallel lines. Van der Waals interactions are shown as grey dashed lines and polar interactions as blue dashed lines. (B) Interaction of XMRV RT with the terminal portion of the RNA/DNA duplex (shown in red and blue for RNA and DNA, respectively). XMRV RT protein is in surface representation. Tyr64 and residues forming the ‘pin’ stabilizing the conformation of the template in front of the incoming nucleotide are shown as cyan sticks, and the dNTP modelled based on the HIV-1 RT structure (PDB ID: 1RTD) is shown as dark grey sticks. (C) Interactions with the 2′-OH groups of the template. The RNA strand is shown as red sticks and 2′-OH groups as spheres. Hydrogen bonds are indicated with dashed lines. (D) Binding of primer nucleotides by residues of the thumb subdomain. In panels (B) and (D), a composite simulated annealing omit map contoured at 1 σ is overlaid on the substrate (blue mesh).