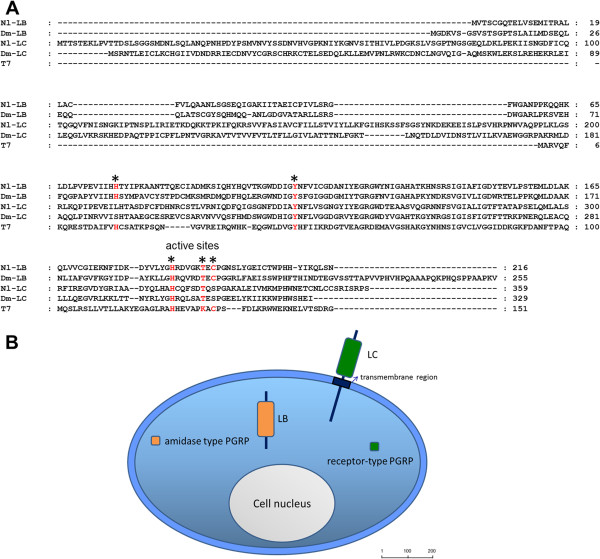
Figure 1.
(A) Multiple alignments of PGRPs and T7 lysozyme. The ClustalX program was used for alignments. The GenBank accession numbers for the sequences are as follows: N. lugens PGRP-LB (KC355211); N. lugens PGRP-LC (KC355212); D. melanogaster PGRP-LB (AFH06370); D. melanogaster PGRP-LC (ACZ94668) and Enterobacteria phage T7 lysozyme (AAB32819). Five amino acid residues required for amidase activity are marked by asterisks and shown in red. (B) Predicted cellular distribution of N. lugens PGRPs. N. lugens PGRP-LC is likely a receptor protein due to its transmembrane region. PGRP-LB lacks the signal peptide and transmembrane region, thus possibly making it a cytosolic protein. The potentially catalytic or non-catalytic amidase activity of the PGRP proteins is shown in orange and green respectively. The size bar indicates the amino acid residues of the deduced proteins.