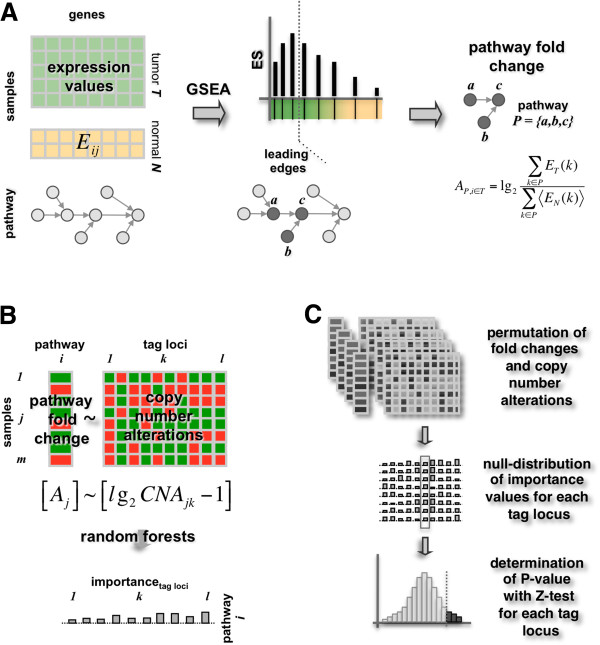
Figure 1.
Outline of the procedure. (A) We used 158 gene expression samples of Glioblastoma Multiforme patients (GBM) and 21 non-tumor control samples. Using 181 pathways we applied Gene Set Enrichment Analysis (GSEA) and found 119 over- and 62 under-expressed pathways. Determining leading edge genes that govern their over/under expression when comparing disease to control cases we represented each pathway by its expression fold change score. (B) We fitted pathway fold change scores as a function of the corresponding copy number variations of tag-loci. Using random forest algorithm we obtained a normalized importance score of each locus/pathway pair. (C) To assess the statistical significance of a tag-locus’ importance for fitting a pathways expression we performed permutation tests by randomizing both pathway fold change scores and copy number alterations. Focusing on such random distributions of importance scores we applied a Z-test to determine P-values, allowing us to assess the significance of an association between each tag-locus and pathway.