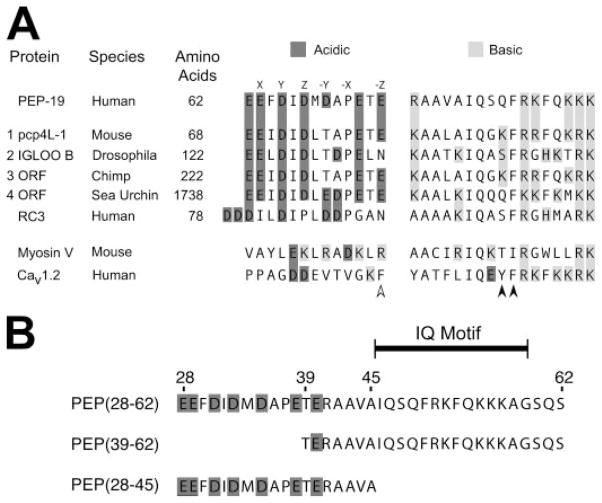
FIGURE 1. Sequence alignment of PEP-19 with residues in other proteins.
A protein BLAST similarity search was performed using the NCBI web server to compare PEP-19 residues 25–62 with all non-redundant sequences in GenBank CDS translations, RefSeq proteins, PDB, SwissProt, PIR, and PRF databases. Panel A shows the sequence similarity between PEP-19 and 4 of 149 BLAST hits. Also shown are the corresponding regions from RC3, myosin V, and voltage-gated Ca2+ channel (Cav1.2). The letters above residues in the PEP-19 sequence are the relative positions of amino acids that coordinate Ca2+ in a consensus EF-hand (see text for details). The accession numbers for BLAST hits 1 to 4 are: AY304481.1, NP_477061, XM_001152960.1, and XM_787004.2. Panel B shows sequences for PEP-19 synthetic peptides used in the current study. The indicated IQ motif is based on a comparison of numerous IQ motif proteins (10), and the minimal region in PEP-19 that has CaM antagonist activity (36).