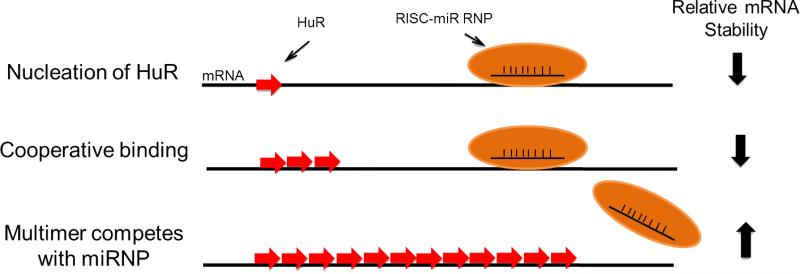
Figure 2. Ribonucleosome model depicting antagonism of the miR/RISC mRNA destabilizing complex by proximal binding of HuR and possibly other members of the ELAV/Hu RBP family [27,82].
In the example, a single molecule of HuR binds at a U-rich RNA-recognition element followed by cooperative binding of multiple HuR molecules [15,19-21,31-37]. Depending on the exact spacing between the nucleation site and the miR/RISC, multimeric binding of HuR could result in steric hindrance or physical displacement of the RISC. This event would logically antagonize the destabilization functions of miR/RISC [29] resulting in increased stability of the mRNA target by HuR, and plausibly, all ELAV/Hu RBPs as demonstrated [16-17,21-23]. The model suggests that a HuR ribonucleosome may displace the miRNP to stabilize mRNA targets. However, the precise binding motifs and coding rules that determine nucleation and multimerization of HuR are poorly understood and may differ with each message [15,31,34,35].