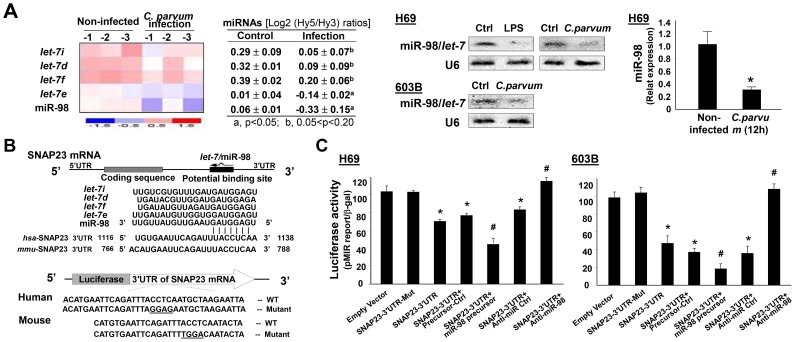
Figure 5. The let-7 miRNAs target SNAP23 and are downregulated in epithelial cells following C. parvum infection.
(A) C. parvum infection downregulates expression of the let-7 miRNA family. H69 cells were exposed to C. parvum infection for 12 h, followed by miRNA analysis using mercury LNA array, Northern blot, and real-time PCR. A heat map of the let-7 miRNA family is shown and expression levels are presented as the log2 (Hy5/Hy3) ratios. Representative Northern blot for let-7/miR-98 in H69 and 603B cells and real-time PCR quantification of miR-98 in H69 cells are shown. (B) The schematic of SNAP23 mRNA showed a potential binding site in its 3′UTR for the let-7 miRNA family in humans and in mice. The SNAP23 3′UTR sequence covering the potential binding sites for the let-7 miRNA family was inserted into the pMIR-REPORT luciferase plasmid. A control plasmid with the mutant 3′UTR sequence was also generated for control. (C) Binding of let-7/miR-98 miRNAs to the potential binding site in the SNAP23 3′UTR results in translational suppression. Cells were transfected with the pMIR-REPORTER luciferase constructs and treated with anti-miRs or precursors to miR-98, or non-specific oligo control, for 24 h, followed by luciferase analysis. *, p<0.05 ANOVA versus the non-infected control (in A) or empty vector control (in C); #, p<0.05 ANOVA versus the control precursor or control anti-miR (in C).