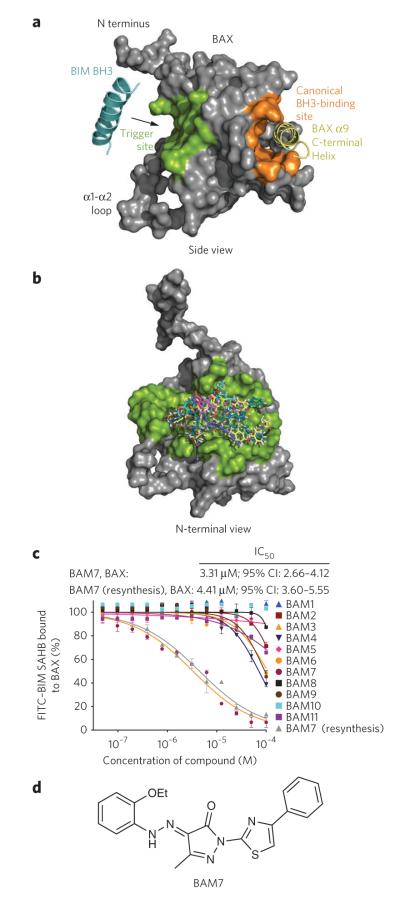
Figure 1. In silico screen for small-molecule binders of the BAX trigger site identifies BAM7.
(a) The BIM BH3 trigger site localizes to the N-terminal face of BAX, as highlighted in green in this side view of the protein. In contrast, the canonical BH3 binding pocket of antiapoptotic proteins (orange) maps to the opposite side of BAX and remains occupied by the C-terminal helix 9 (yellow) when the protein is in the inactive, monomeric state. (b) A computational screening algorithm using an in silico library of 750,000 small molecules docked on average minimized BAX structures yielded a panel of 100 candidate BAMs. A compilation of the docked structures demonstrates how candidate BAMs occupy the topographic landscape of the BAX trigger site (green). (c) BAM7 emerged as the most effective small-molecule binder (IC50, 3.3 μM) in a competitive FPA involving FITC-BIM SAHB and BAX. The structural identity of BAM7 was confirmed by NMR and MS, and the molecule was resynthesized and found to have a similar IC50 (4.4 μM) upon repeat testing by competitive FPA. 95% CI, 95% confidence interval. Data are mean and s.d. for experiments performed in at least triplicate. (d) BAM7 is a 405.5-Da small molecule whose chemical features include a pyrazolone core substituted with ethoxyphenylhydrazono, methyl and phenylthiazol R groups.