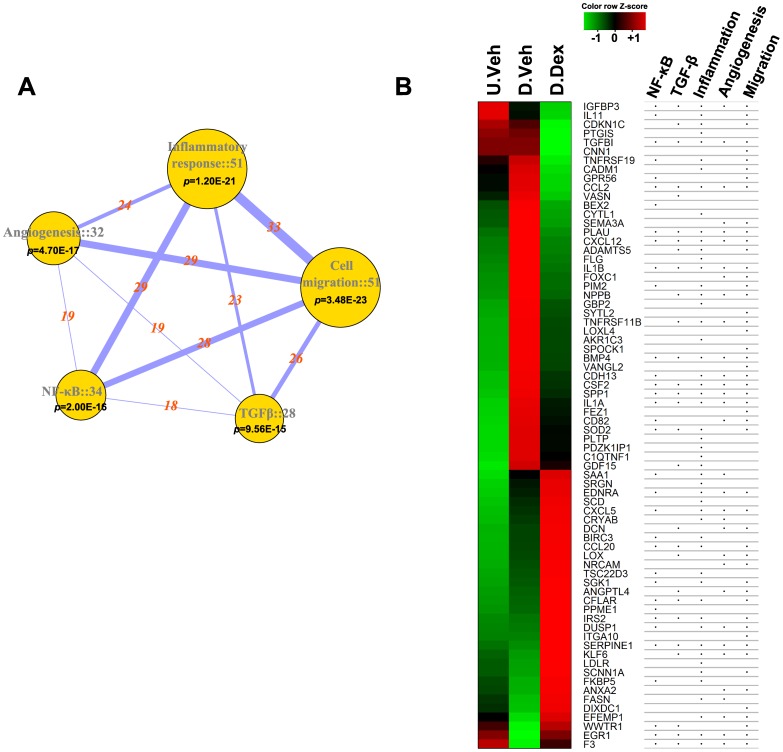
Figure 6. Entrez keyword-based functional annotation analysis unveils unique effects of Dex treatment during HPC differentiation.
The most significantly changed probes (>2 folds and q<0.01) were used for the analysis. The Entrez keywords tested were: Inflammatory response, Cell migration, Angiogenesis, NF-κB and TGFβ. The gene networks associated with these keywords were retrieved from NCBI Entrez database [29]. (A) The gene networks of the tested keywords are summarized. The number of significantly changed genes and the p values are shown with the category nodes. The node size is proportional to gene number. The numbers of common genes shared by different category nodes are marked with the connecting lines. The width of line is proportional to the number. (B) Dex effects on the gene expression during HPC differentiation. The genes were annotated by the different functional categories.