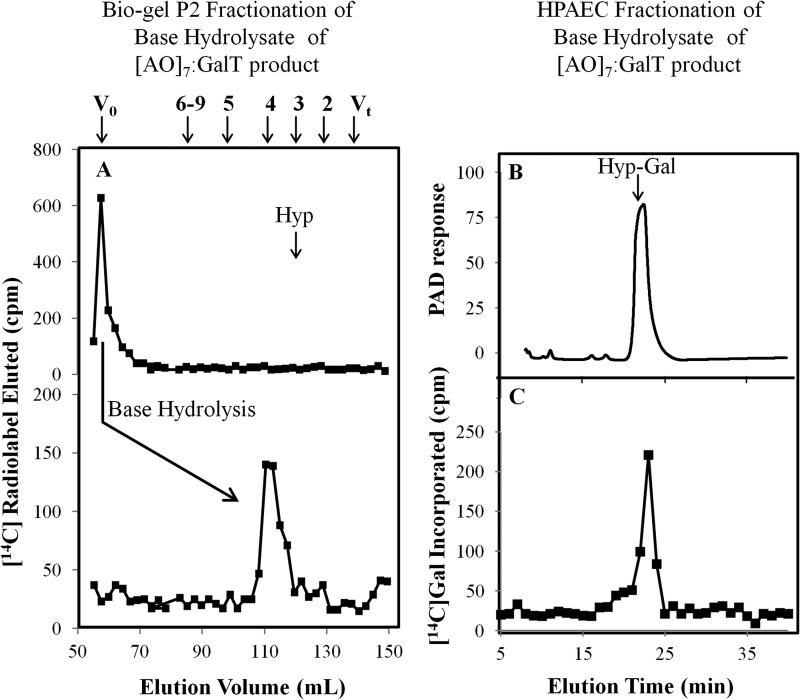
FIGURE 4.
Bio-gel P2 fractionation of the RP-HPLC-purified (AO)7:AtGALT2 reaction product and HPAEC of the resulting base hydrolysis product. A, Bio-gel P2 fractionation of the RP-HPLC-purified (AO)7:AtGALT2 reaction product before and after base hydrolysis. Permeabilized microsomal membranes from the Pichia C2 line expressing His6-GALT2 served as the enzyme source in the (AO)7:AtGALT2 reaction. Elution profiles of the reaction product before and after base hydrolysis are shown. The column was calibrated with high molecular mass dextran (V0), galactose (Vt), xylo-oligosaccharides with DP2 to -5, and xyloglucan-oligosaccharides (DP6 to -9); their elution positions are indicated with arrows at the top. The elution position of free Hyp amino acid (corresponding to DP3) is shown with an arrow in the panel. Base hydrolysis produces a radioactive peak eluting at DP4, which corresponds to Hyp-Gal. B, HPAEC profile of a chemically synthesized Hyp-Gal standard detected as a PAD response. C, the radioactive peak eluting at DP4 coelutes with the chemically synthesized Hyp-Gal standard following HPAEC. Both the Hyp-Gal standard and the radioactive peak eluting at DP4 were fractionated in 20 mm NaOH elution buffer on a CarboPac PA-20 column.