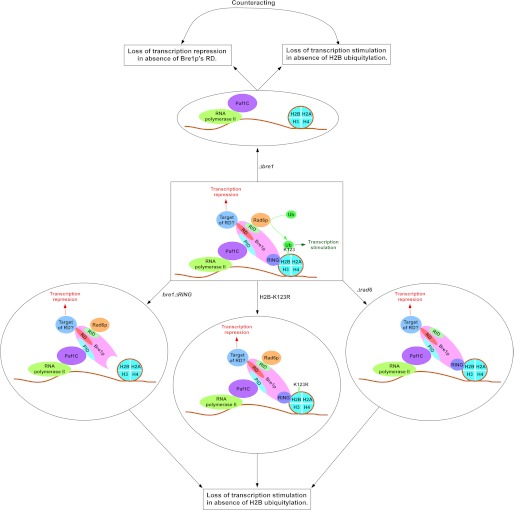
FIGURE 8.
The schematic diagram showing the stimulatory and repressive roles of Bre1p in transcription. Bre1p interacts with chromatin via its RING domain at the C-terminal (19). Rad6p interacts with a domain within the first 210 amino acids at the N-terminal (19), and leads to targeted histone H2B ubiquitylation (19). Paf1C interacts with Bre1p (19) via its non-RING domain to promote histone H2B ubiquitylation activity of Rad6p-Bre1p (19, 34). Thus, bre1ΔRING, Δpaf1, Δrad6, and H2B-K123R mutant strains impair histone H2B ubiquitylation (9, 15–17, 19), RNA polymerase II association, and transcription (8–12; this study). The non-RING part of Bre1p has a repression domain (RD) that lowers the association of RNA polymerase II with the active gene and hence transcription (this study), possibly via interaction with an unknown factor. The transcription stimulation is lost in the absence of histone H2B ubiquitylation in the bre1ΔRING, Δrad6, and H2B-K123R mutant strains. When the whole BRE1 is deleted, transcription repression is lost in the absence of the repression domain of Bre1p, in addition to impairment of transcriptional stimulation. These two opposing activities counteract, and hence the defect in transcription (or RNA polymerase II association) is not apparently observed in the Δbre1 strain. The bre1Δ50 mutant is represented by bre1ΔRING (i.e. Bre1p without RING domain). PID, Paf1C interaction domain; RID, Rad6p interaction domain; Ub, ubiquitin.